- Privacy Policy

Home » Prediction – Definition, Types and Example

Prediction – Definition, Types and Example
Table of Contents

Definition:
Prediction is the process of making an educated guess or estimation about a future event or outcome based on available information and data. It involves analyzing past patterns and trends, as well as current conditions, to forecast what may happen in the future.
Types of Prediction
Types of Prediction are as follows:
Point Prediction
This type of prediction provides a specific estimate of the future outcome. For example, predicting that a stock will reach a certain price at a particular time.
Interval Prediction
This type of prediction provides a range of possible outcomes. For example, predicting that there is a 90% chance that a hurricane will make landfall somewhere in a certain region within the next week.
Categorical Prediction
This type of prediction involves predicting the likelihood of an event occurring in a specific category. For example, predicting the likelihood of a person developing a certain disease or the likelihood of a sports team winning a game.
Long-term Prediction
This type of prediction involves forecasting events or trends that are expected to occur over a longer period, such as predicting climate change or population growth.
Short-term Prediction
This type of prediction involves forecasting events or trends that are expected to occur within a shorter period, such as predicting the weather or the stock market performance for the next day.
Qualitative Prediction
This type of prediction involves making subjective judgments or expert opinions based on non-quantifiable information, such as predicting the impact of a new technology on society.
Quantitative Prediction
This type of prediction involves using mathematical models and statistical methods to forecast future events or trends, such as predicting consumer demand for a new product.
Probabilistic Prediction
This type of prediction involves estimating the probability or likelihood of a future event or outcome occurring. For example, predicting the probability of a person surviving a medical procedure.
Deterministic Prediction
This type of prediction involves providing a definite or certain outcome based on known information. For example, predicting the result of a coin toss or the outcome of a mathematical equation.
Black box Prediction
This type of prediction involves using machine learning algorithms or other complex models to make predictions without necessarily understanding how the model arrived at its conclusion. This type of prediction is often used in applications such as fraud detection or image recognition.
Prediction Methods
Prediction Methods are as follows:
Statistical Methods
These methods involve analyzing historical data and using statistical models to identify patterns and trends that can be used to make predictions about future events or outcomes.
Machine Learning Methods
These methods involve training algorithms to learn patterns and relationships in data and using these models to make predictions about new data.
Expert Judgment
This method involves relying on the knowledge and expertise of individuals who have specialized knowledge in a particular area to make predictions.
Simulation Methods
These methods involve creating computer models that simulate real-world situations to predict outcomes. For example, simulating the spread of a virus in a population to predict the impact of different intervention strategies.
Rule-based Methods
These methods involve using a set of rules or decision trees to make predictions based on specific criteria. For example, using a set of rules to predict the likelihood of a loan being approved based on a person’s credit history and income.
Time-series Forecasting
This method involves analyzing historical data to identify patterns and trends over time and using these patterns to make predictions about future values in a series, such as predicting stock prices or demand for a product.
Neural Networks
These are a type of machine learning method that involve building networks of interconnected nodes that can learn to make predictions based on input data.
Examples of Prediction
There are numerous examples of predictions made in various fields, some of which include:
- Weather forecasting: Predicting the temperature, precipitation, and other weather conditions for a particular location and time.
- Stock market prediction : Predicting the performance of stocks, bonds, and other financial instruments based on market trends and other economic factors.
- Sports prediction : Predicting the outcomes of sporting events such as football games, horse races, and tennis matches.
- Healthcare prediction : Predicting the likelihood of a patient developing a particular disease or the effectiveness of a particular treatment.
- Natural disaster prediction : Predicting the occurrence and intensity of natural disasters such as hurricanes, earthquakes, and floods.
- Traffic prediction: Predicting traffic patterns and congestion in urban areas based on historical data and other factors.
- Retail prediction: Predicting consumer demand for products and services based on market trends, customer behavior, and other factors.
- Energy prediction : Predicting energy demand and supply based on historical data, weather patterns, and other factors.
Applications of Prediction
Predictive models and methods have numerous applications across a wide range of fields, some of which include:
- Business and finance: Predicting sales, customer behavior, and market trends to inform business planning and decision-making, and predicting stock prices and other financial market performance.
- Healthcare : Predicting disease diagnosis, treatment outcomes, and drug efficacy to inform patient care and medical research.
- Weather forecasting: Predicting weather patterns and conditions to inform emergency response planning, agriculture, and transportation.
- Transportation : Predicting traffic patterns and congestion to inform route planning and transportation infrastructure development.
- Sports : Predicting the outcomes of sporting events to inform sports betting and game strategy.
- Marketing : Predicting consumer behavior, preferences, and buying habits to inform marketing and advertising strategies.
- Education : Predicting student performance and outcomes to inform academic planning and intervention strategies.
- Energy and utilities : Predicting energy demand and supply to inform energy infrastructure planning and maintenance.
Purpose of Prediction
The purpose of prediction is to make informed decisions and take actions based on expected future outcomes. Predictions are used to estimate the likelihood of future events or outcomes, and to guide decision-making based on those estimates.
In many industries and fields, predictions are an essential tool for optimizing resources, managing risks, and improving outcomes. For example, in finance, stock market predictions are used to inform investment decisions, and in healthcare, disease prediction models are used to identify patients at risk of developing certain conditions and inform treatment decisions.
Predictions are also used to anticipate and prepare for potential future events or outcomes, such as natural disasters, epidemics, or economic downturns. By using predictions to prepare for these scenarios, businesses, governments, and organizations can reduce the impact of such events and improve their resilience.
When to Predict
Here are some common situations where predictions are made:
- Before making a decision : Predictions can be made before making a decision to inform the decision-making process. For example, predicting sales or market trends before launching a new product to help inform marketing and pricing decisions.
- During planning and forecasting : Predictions can be made during planning and forecasting processes to inform resource allocation and strategy development. For example, predicting demand for products or services to inform production and supply chain planning.
- In response to emerging situations: Predictions can be made in response to emerging situations, such as natural disasters, pandemics, or economic changes. For example, predicting the spread of a virus to inform public health interventions.
- To improve performance: Predictions can be made to identify areas for improvement and to optimize performance. For example, predicting equipment failures to inform maintenance schedules and reduce downtime.
Advantages of Prediction
Some of the advantages of prediction include:
- Improved decision-making : Predictions provide valuable insights into future outcomes, helping decision-makers to make more informed and effective decisions.
- Risk management: Predictions can help identify and manage risks by providing estimates of the likelihood and potential impact of future events.
- Resource optimization : Predictions can inform resource allocation and optimization, allowing businesses and organizations to use their resources more efficiently and effectively.
- Cost savings: Predictions can help identify opportunities to reduce costs and increase efficiency by identifying areas for improvement.
- Competitive advantage : Predictions can give businesses and organizations a competitive advantage by enabling them to anticipate market trends and respond quickly to changes.
- Improved outcomes: Predictions can lead to improved outcomes, whether in healthcare, finance, or other fields, by helping to identify high-risk individuals or optimizing treatment plans.
- Planning and forecasting: Predictions can inform planning and forecasting processes, enabling businesses and organizations to anticipate and prepare for future events and outcomes.
Disadvantages of Prediction
Here are some of the main disadvantages of prediction:
- Uncertainty : Predictions are inherently uncertain and are based on assumptions and data that may not always be accurate or complete. This can lead to errors and inaccuracies in the prediction.
- Over-reliance on predictions: Over-reliance on predictions can lead to complacency and a failure to consider other important factors or to adapt to changing circumstances.
- Ethical concerns : Predictions can raise ethical concerns, particularly when they involve sensitive topics such as healthcare or criminal justice. For example, using predictions to make decisions about medical treatment or criminal sentencing may be seen as unfair or discriminatory.
- Limited data availability : Predictions are only as good as the data that is available to support them. In some cases, there may be limited or incomplete data available, which can make it difficult to develop accurate predictions.
- Bias : Predictions may be biased if the data used to develop them is biased or if the algorithms used to generate the predictions have inherent biases.
- Unforeseen events : Predictions may not account for unforeseen events that can impact the outcome being predicted. For example, a natural disaster or other unexpected event could significantly alter the outcome being predicted.
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

What is Art – Definition, Types, Examples

What is Anthropology – Definition and Overview

What is Literature – Definition, Types, Examples

Economist – Definition, Types, Work Area

Anthropologist – Definition, Types, Work Area

What is History – Definitions, Periods, Methods
- - Google Chrome
Intended for healthcare professionals
- Access provided by Google Indexer
- My email alerts
- BMA member login
- Username * Password * Forgot your log in details? Need to activate BMA Member Log In Log in via OpenAthens Log in via your institution

Search form
- Advanced search
- Search responses
- Search blogs
- A guide to systematic...
A guide to systematic review and meta-analysis of prediction model performance
- Related content
- Peer review
- Thomas P A Debray , assistant professor 1 2 ,
- Johanna A A G Damen , PhD fellow 1 2 ,
- Kym I E Snell , research fellow 3 ,
- Joie Ensor , research fellow 3 ,
- Lotty Hooft , associate professor 1 2 ,
- Johannes B Reitsma , associate professor 1 2 ,
- Richard D Riley , professor 3 ,
- Karel G M Moons , professor 1 2
- 1 Cochrane Netherlands, University Medical Center Utrecht, PO Box 85500 Str 6.131, 3508 GA Utrecht, Netherlands
- 2 Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, PO Box 85500 Str 6.131, 3508 GA Utrecht, Netherlands
- 3 Research Institute for Primary Care and Health Sciences, Keele University, Staffordshire, UK
- Correspondence to: T P A Debray T.Debray{at}umcutrecht.nl
- Accepted 25 November 2016
Validation of prediction models is highly recommended and increasingly common in the literature. A systematic review of validation studies is therefore helpful, with meta-analysis needed to summarise the predictive performance of the model being validated across different settings and populations. This article provides guidance for researchers systematically reviewing and meta-analysing the existing evidence on a specific prediction model, discusses good practice when quantitatively summarising the predictive performance of the model across studies, and provides recommendations for interpreting meta-analysis estimates of model performance. We present key steps of the meta-analysis and illustrate each step in an example review, by summarising the discrimination and calibration performance of the EuroSCORE for predicting operative mortality in patients undergoing coronary artery bypass grafting.
Summary points
Systematic review of the validation studies of a prediction model might help to identify whether its predictions are sufficiently accurate across different settings and populations
Efforts should be made to restore missing information from validation studies and to harmonise the extracted performance statistics
Heterogeneity should be expected when summarising estimates of a model’s predictive performance
Meta-analysis should primarily be used to investigate variation across validation study results
Systematic reviews and meta-analysis are an important—if not the most important—source of information for evidence based medicine. 1 Traditionally, they aim to summarise the results of publications or reports of primary treatment studies and (more recently) of primary diagnostic test accuracy studies. Compared to therapeutic intervention and diagnostic test accuracy studies, there is limited guidance on the conduct of systematic reviews and meta-analysis of primary prognosis studies.
A common aim of primary prognostic studies concerns the development of so-called prognostic prediction models or indices. These models estimate the individualised probability or risk that a certain condition will occur in the future by combining information from multiple prognostic factors from an individual. Unfortunately, there is often conflicting evidence about the predictive performance of developed prognostic prediction models. For this reason, there is a growing demand for evidence synthesis of (external validation) studies assessing a model’s performance in new individuals. 2 A similar issue relates to diagnostic prediction models, where the validation performance of a model for predicting the risk of a disease being already present is of interest across multiple studies.
Previous guidance papers regarding methods for systematic reviews of predictive modelling studies have addressed the searching, 3 4 5 design, 2 data extraction, and critical appraisal 6 7 of primary studies. In this paper, we provide further guidance for systematic review and for meta-analysis of such models. Systematically reviewing the predictive performance of one or more prediction models is crucial to examine a model’s predictive ability across different study populations, settings, or locations, 8 9 10 11 and to evaluate the need for further adjustments or improvements of a model.
Although systematic reviews of prediction modelling studies are increasingly common, 12 13 14 15 16 17 researchers often refrain from undertaking a quantitative synthesis or meta-analysis of the predictive performance of a specific model. Potential reasons for this pitfall are concerns about the quality of included studies, unavailability of relevant summary statistics due to incomplete reporting, 18 or simply a lack of methodological guidance.
Based on previous publications, we therefore first describe how to define the systematic review question, to identify the relevant prediction modelling studies from the literature 3 5 and to critically appraise the identified studies. 6 7 Additionally, and not yet addressed in previous publications, we provide guidance on which predictive performance measures could be extracted from the primary studies, why they are important, and how to deal with situations when they are missing or poorly reported. The need to extract aggregate results and information from published studies provides unique challenges that are not faced when individual participant data are available, as described recently in The BMJ . 19
We subsequently discuss how to quantitatively summarise the extracted predictive performance estimates and investigate sources of between-study heterogeneity. The different steps are summarised in figure 1 ⇓ , some of which are explained further in different appendices. We illustrate each step of the review using an empirical example study—that is, the synthesis of studies validating predictive performance of the additive European system for cardiac operative risk evaluation (EuroSCORE). Here onwards, we focus on systematic review and meta-analysis of a specific prognostic prediction model. All guidance can, however, similarly be applied to the meta-analysis of diagnostic prediction models. We focus on statistical criteria of good performance (eg, in terms of discrimination and calibration) and highlight other clinically important measures of performance (such as net benefit) in the discussion.
Fig 1 Flowchart for systematically reviewing and, if considered appropriate, meta-analysis of the validation studies of a prediction model. CHARMS=checklist for critical appraisal and data extraction for systematic reviews of prediction modelling studies; PROBAST=prediction model risk of bias assessment tool; PICOTS=population, intervention, comparator, outcome(s), timing, setting; GRADE=grades of recommendation, assessment, development, and evaluation; PRISMA=preferred reporting items for systematic reviews and meta-analyses; TRIPOD=transparent reporting of a multivariable prediction model for individual prognosis or diagnosis
- Download figure
- Open in new tab
- Download powerpoint
Empirical example
As mentioned earlier, we illustrate our guidance using a published review of studies validating EuroSCORE. 13 This prognostic model aims to predict 30 day mortality in patients undergoing any type of cardiac surgery (appendix 1). It was developed by a European steering group in 1999 using logistic regression in a dataset from 13 302 adult patients undergoing cardiac surgery under cardiopulmonary bypass. The previous review identified 67 articles assessing the performance of the EuroSCORE in patients that were not used for the development of the model (external validation studies). 13 It is important to evaluate whether the predictive performance of EuroSCORE is adequate, because poor performance could eventually lead to poor decision making and thereby affect patient health.
In this paper, we focus on the validation studies that examined the predictive performance of the so-called additive EuroSCORE system in patients undergoing (only) coronary artery bypass grafting (CABG). We included a total of 22 validations, including more than 100 000 patients from 20 external validation studies and from the original development study (appendix 2).
Steps of the systematic review
Formulating the review question and protocol.
As for any other type of biomedical research, it is strongly recommended to start with a study protocol describing the rationale, objectives, design, methodology, and statistical considerations of the systematic review. 20 Guidance for formulating a review question for systematic review of prediction models has recently been provided by the CHARMS checklist (checklist for critical appraisal and data extraction for systematic reviews of prediction modelling studies). 6 This checklist addresses a modification (PICOTS) of the PICO system (population, intervention, comparison, and outcome) used in therapeutic studies, and additionally considers timing (that is, at which time point and over what time period the outcome is predicted) and setting (that is, the role or setting of the prognostic model). More information on the different items is provided in box 1 and appendix 3.
Box 1: PICOTS system
The PICOTS system, as presented in the CHARMS checklist, 6 describes key items for framing the review aim, search strategy, and study inclusion and exclusion criteria. The items are explained below in brief, and applied to our case study:
Population—define the target population in which the prediction model will be used. In our case study, the population of interest comprises patients undergoing coronary artery bypass grafting.
Intervention (model)—define the prediction model(s) under review. In the case study, the focus is on the prognostic additive EuroSCORE model.
Comparator—if applicable, one can address competing models for the prognostic model under review. The existence of alternative models was not considered in our case study.
Outcome(s)—define the outcome(s) of interest for which the model is validated. In our case study, the outcome was defined as all cause mortality. Papers validating the EuroSCORE model to predict other outcomes such as cardiovascular mortality were excluded.
Timing—specifically for prognostic models, it is important to define when and over what time period the outcome is predicted. Here, we focus on all cause mortality at 30 days, predicted using preoperative conditions.
Setting—define the intended role or setting of the prognostic model. In the case study, the intended use of the EuroSCORE model was to perform risk stratification in the assessment of cardiac surgical results, such that operative mortality could be used as a valid measure of quality of care.
The formal review question was as follows: to what extent is the additive EuroSCORE able to predict all cause mortality at 30 days in patients undergoing CABG? The question is primarily interested in the predictive performance of the original EuroSCORE, and not how it performs after it has been recalibrated or adjusted in new data.
Formulating the search strategy
When reviewing studies that evaluate the predictive performance of a specific prognostic model, it is important to ensure that the search strategy identifies all publications that validated the model for the target population, setting, or outcomes at interest. To this end, the search strategy should be formulated according to aforementioned PICOTS of interest. Often, the yield of search strategies can further be improved by making use of existing filters for identifying prediction modelling studies 3 4 5 or by adding the name or acronym of the model under review. Finally, it might help to inspect studies that cite the original publication in which the model was developed. 15
We used a generic search strategy including the terms “EuroSCORE” and “Euro SCORE” in the title and abstract. The search resulted in 686 articles. Finally, we performed a cross reference check in the retrieved articles, and identified one additional validation study of the additive EuroSCORE.
Critical appraisal
The quality of any meta-analysis of a systematic review strongly depends on the relevance and methodological quality of included studies. For this reason, it is important to evaluate their congruence with the review question, and to assess flaws in the design, conduct, and analysis of each validation study. This practice is also recommended by Cochrane, and can be implemented using the CHARMS checklist, 6 and, in the near future, using the prediction model risk of bias assessment tool (PROBAST). 7
Using the CHARMS checklist and a preliminary version of the PROBAST tool, we critically appraised the risk of bias of each retrieved validation study of the EuroSCORE, as well as of the model development study. Most (n=14) of the 22 validation studies were of low or unclear risk of bias (fig 2 ⇓ ). Unfortunately, several validation studies did not report how missing data were handled (n=13) or performed complete case analysis (n=5). We planned a sensitivity analysis that excluded all validation studies with high risk of bias for at least one domain (n=8). 21
Fig 2 Overall judgment for risk of bias of included articles in the case study (predictive performance of the EuroSCORE for all cause mortality at 30 days in patients undergoing coronary artery bypass grafting). Study references listed in appendix 2. Study participants domain=design of the included validation study, and inclusion and exclusion of its participants; predictors domain=definition, timing, and measurement of predictors in the validation study (it also assesses whether predictors have not been measured and were therefore omitted from the model in the validation study); outcome domain=definition, timing, and measurement of predicted outcomes; sample size and missing data domain=number of participants in the validation study and exclusions owing to missing data; statistical analysis domain=validation methods (eg, whether the model was recalibrated before validation). Note that there are two validations presented in Nashef 2002; the same scores apply to both model validations. *Original development study (split sample validation)
Quantitative data extraction and preparation
To allow for quantitative synthesis of the predictive performance of the prediction model under study, the necessary results or performance measures and their precision need to be extracted from each model validation study report. The CHARMS checklist can be used for this guidance. We briefly highlight the two most common statistical measures of predictive performance, discrimination and calibration, and discuss how to deal with unreported or inconsistent reporting of these performance measures.
Discrimination
Discrimination refers to a prediction model’s ability to distinguish between patients developing and not developing the outcome, and is often quantified by the concordance (C) statistic. The C statistic ranges from 0.5 (no discriminative ability) to 1 (perfect discriminative ability). Concordance is most familiar from logistic regression models, where it is also known as the area under the receiver operating characteristics (ROC) curve. Although C statistics are the most common reported estimates of prediction model performance, they can still be estimated from other reported quantities when missing. Formulas for doing this are presented in appendix 7 (along with their standard errors), and implement the transformations that are needed for conducting the meta-analysis (see meta-analysis section below).
The C statistic of a prediction model can vary substantially across different validation studies. A common cause for heterogeneity in reported C statistics relates to differences between studied populations or study designs. 8 22 In particular, it has been demonstrated that the distribution of patient characteristics (so-called case mix variation) could substantially affect the discrimination of the prediction model, even when the effects of all predictors (that is, regression coefficients) remain correct in the validation study. 22 The more similarity that exists between participants of a validation study (that is, a more homogeneous or narrower case mix), the less discrimination can be achieved by the prediction model.
Therefore, it is important to extract information on the case mix variation between patients for each included validation study, 8 such as the standard deviation of the key characteristics of patients, or of the linear predictor (fig 3 ⇓ ). The linear predictor is the weighted sum of the values of the predictors in the validation study, where the weights are the regression coefficients of the prediction model under investigation. 23 Heterogeneity in reported C statistics might also appear when predictor effects differ across studies (eg, due to different measurement methods of predictors), or when different definitions (or different derivations) of the C statistic have been used. Recently, several concordance measures have been proposed that allow to disentangle between different sources of heterogeneity. 22 24 Unfortunately, these measures are currently rarely reported.
Fig 3 Estimation of the standard deviation of the linear predictor as a way to quantify case mix variation within a study
We found that the C statistic of the EuroSCORE was reported in 20 validations (table 1 ⇓ ). When measures of uncertainty were not reported, we approximated the standard error of the C statistic (seven studies) using the equations provided in appendix 7 (fig 4 ⇓ ). Furthermore, for each validation, we extracted the standard deviation of the age distribution and of the linear predictor of the additive EuroSCORE to help quantify the case mix variation in each study. When such information could not be retrieved, we estimated the standard deviation from reported ranges or histograms (fig 3 ⇑ ). 26
Details of the 22 validations of the additive EuroSCORE to predict overall mortality at 30 days
- View inline
Fig 4 Forest plots of extracted performance statistics of the additive EuroSCORE in the case study (to predict all cause mortality at 30 days in patients undergoing coronary artery bypass grafting). Part A shows forest plot of study specific C statistics (all 95% confidence intervals estimated on the logit scale); part B shows forest plot of study specific total O:E ratios (where O=total number of observed deaths and E=total number of expected deaths as predicted by the model; when missing, 95% confidence intervals were approximated on the log scale using the equations from appendix 7). *Performance in the original development study (split sample validation)
Calibration
Calibration refers to a model’s accuracy of predicted risk probabilities, and indicates the extent to which expected outcomes (predicted from the model) and observed outcomes agree. It is preferably reported graphically with expected outcome probabilities plotted against observed outcome frequencies (so-called calibration plots, see appendix 4), often across tenths of predicted risk. 23 Also for calibration, reported performance estimates might vary across different validation studies. Common causes for this are differences in overall prognosis (outcome incidence). These differences might appear because of differences in healthcare quality and delivery, for example, with screening programmes in some countries identifying disease at an earlier stage, and thus apparently improving prognosis in early years compared to other countries. This again emphasises the need to identify studies and participants relevant to the target population, so that a meta-analysis of calibration performance is relevant.
Summarising estimates of calibration performance is challenging because calibration plots are most often not presented, and because studies tend to report different types of summary statistics in calibration. 12 27 Therefore, we propose to extract information on the total number of observed (O) and expected (E) events, which are statistics most likely to be reported or derivable (appendix 7). The total O:E ratio provides a rough indication of the overall model calibration (across the entire range of predicted risks). The total O:E ratio is strongly related to the calibration in the large (appendix 5), but that is rarely reported. The O:E ratio might also be available in subgroups, for example, defined by tenths of predicted risk or by particular groups of interest (eg, ethnic groups, or regions). These O:E ratios could also be extracted, although it is unlikely that all studies will report the same subgroups. Finally, it would be helpful to also extract and summarise estimates of the calibration slope.
Calibration of the additive EuroSCORE was visually assessed in seven validation studies. Although the total O:E ratio was typically not reported, it could be calculated from other information for 19 of the 22 included validations. For nine of these validation studies, it was also possible to extract the proportion of observed outcomes across different risk strata of the additive EuroSCORE (appendix 8). Measures of uncertainty were often not reported (table 1 ⇑ ). We therefore approximated the standard error of the total O:E ratio (19 validation studies) using the equations provided in appendix 7. The forest plot displaying the study specific results is presented in figure 4 ⇑ . The calibration slope was not reported for any validation study and could not be derived using other information.
Performance of survival models
Although we focus on discrimination and calibration measures of prediction models with a binary outcome, similar performance measures exist for prediction models with a survival (time to event) outcome. Caution is, however, warranted when extracting reported C statistics because different adaptations have been proposed for use with time to event outcomes. 9 28 29 We therefore recommend to carefully evaluate the type of reported C statistic and to consider additional measures of model discrimination.
For instance, the D statistic gives the log hazard ratio of a model’s predicted risks dichotomised at the median value, and can be estimated from Harrell’s C statistic when missing. 30 Finally, when summarising the calibration performance of survival models, it is recommended to extract or calculate O:E ratios for particular (same) time points because they are likely to differ across time. When some events remain unobserved, owing to censoring, the total number of events and the observed outcome risk at particular time points should be derived (or approximated) using Kaplan-Meier estimates or Kaplan-Meier curves.
Meta-analysis
Once all relevant studies have been identified and corresponding results have been extracted, the retrieved estimates of model discrimination and calibration can be summarised into a weighted average. Because validation studies typically differ in design, execution, and thus case-mix, variation between their results are unlikely to occur by chance only. 8 22 For this reason, the meta-analysis should usually allow for (rather than ignore) the presence of heterogeneity and aim to produce a summary result (with its 95% confidence interval) that quantifies the average performance across studies. This can be achieved by implementing a random (rather than a fixed) effects meta-analysis model (appendix 9). The meta-analysis then also yields an estimate of the between-study standard deviation, which directly quantifies the extent of heterogeneity across studies. 19 Other meta-analysis models have also been proposed, such as by Pennells and colleagues, who suggest weighting by the number of events in each study because this is the principal determinant of study precision. 31 However, we recommend to use traditional random effects models where the weights are based on the within-study error variance. Although it is common to summarise estimates of model discrimination and calibration separately, they can also jointly be synthesised using multivariate meta-analysis. 9 This might help to increase precision of summary estimates, and to avoid exclusion of studies for which relevant estimates are missing (eg, discrimination is reported but not calibration).
To further interpret the relevance of any between-study heterogeneity, it is also helpful to calculate an approximate 95% prediction interval (appendix 9). This interval provides a range for the potential model performance in a new validation study, although it will usually be very wide if there are fewer than 10 studies. 32 It is also possible to estimate the probability of good performance when the model is applied in practice. 9 This probability can, for instance, indicate the likelihood of achieving a certain C statistic in a new population. In case of multivariate meta-analysis, it is even possible to define multiple criteria of good performance. Unfortunately, when performance estimates substantially vary across studies, summary estimates might not be very informative. Of course, it is also desirable to understand the cause of between-study heterogeneity in model performance, and we return to this issue in the next section.
Some caution is warranted when summarising estimates of model discrimination and calibration. Previous studies have demonstrated that extracted C statistics 33 34 35 and total O:E ratios 33 should be rescaled before meta-analysis to improve the validity of its underlying assumptions. Suggestions for the necessary transformations are provided in appendix 7. Furthermore, in line with previous recommendations, we propose to adopt restricted maximum likelihood (REML) estimation and to use the Hartung-Knapp-Sidik-Jonkman (HKSJ) method when calculating 95% confidence intervals for the average performance, to better account for the uncertainty in the estimated between-study heterogeneity. 36 37 The HKSJ method is implemented in several meta-analysis software packages, including the metareg module in Stata (StataCorp) and the metafor package in R (R Foundation for Statistical Computing).
To summarise the performance of the EuroSCORE, we performed random effects meta- analyses with REML estimation and HKSJ confidence interval derivation. For model discrimination, we found a summary C statistic of 0.79 (95% confidence interval 0.77 to 0.81; approximate 95% prediction interval 0.72 to 0.84). The probability of so-called good discrimination (defined as a C statistic >0.75) was 89%. For model calibration, we found a summary O:E ratio of 0.53. This implies that, on average, the additive EuroSCORE substantially overestimates the risk of all cause mortality at 30 days. The weighted average of the total O:E ratio is, however, not very informative because 95% prediction intervals are rather wide (0.19 to 1.46). This problem is also illustrated by the estimated probability of so-called good calibration (defined as an O:E ratio between 0.8 and 1.2), which was only 15%. When jointly meta-analysing discrimination and calibration performance, we found similar summary estimates for the C statistic and total O:E ratio. The joint probability of good performance (defined as C statistic >0.75 and O:E ratio between 0.8 and 1.2), however, decreased to 13% owing to the large extent of miscalibration. Therefore, it is important to investigate potential sources of heterogeneity in the calibration performance of the additive EuroSCORE model.
Investigating heterogeneity across studies
When the discrimination or calibration performance of a prediction model is heterogeneous across validation studies, it is important to investigate potential sources of heterogeneity. This may help to understand under what circumstances the model performance remains adequate, and when the model might require further improvements. As mentioned earlier, the discrimination and calibration of a prediction model can be affected by differences in the design 38 and in populations across the validation studies, for example, owing to changes in case mix variation or baseline risk. 8 22
In general, sources of heterogeneity can be explored by performing a meta-regression analysis where the dependent variable is the (transformed) estimate of the model performance measure. 39 Study level or summarised patient level characteristics (eg, mean age) are then used as explanatory or independent variables. Alternatively, it is possible to summarise model performance across different clinically relevant subgroups. This approach is also known as subgroup analysis and is most sensible when there are clearly definable subgroups. This is often only practical if individual participant data are available. 19
Key issues that could be considered as modifiers of model performance are differences in the heterogeneity between patients across the included validation studies (difference case mix variation), 8 differences in study characteristics (eg, in terms of design, follow-up time, or outcome definition), and differences in the statistical analysis or characteristics related to selective reporting and publication (eg, risk of bias, study size). The regression coefficient obtained from a meta-regression analysis describes how the dependent variable (here, the logit C statistic or log O:E ratio) changes between subgroups of studies in case of a categorical explanatory variable or with one unit increase in a continuous explanatory variable. The statistical significance measure of the regression coefficient is a test of whether there is a (linear) relation between the model’s performance and the explanatory variable. However, unless the number of studies is reasonably large (>10), the power to detect a genuine association with these tests will usually be low. In addition, it is well known that meta-regression and subgroup analysis are prone to ecological bias when investigating summarised patient level covariates as modifiers of model performance. 40
To investigate whether population differences generated heterogeneity across the included validation studies, we performed several meta-regression analyses (fig 5 ⇓ and appendix 10). We first evaluated whether the summary C statistic was related to the case mix variation, as quantified by the spread of the EuroSCORE in each validation study, or related to the spread of patient age. We then evaluated whether the summarised O:E ratio was related to the mean EuroSCORE values, year of study recruitment, or continent. Although the power was limited to detect any association, results suggest that the EuroSCORE tends to overestimate the risk of early mortality in low risk populations (with a mean EuroSCORE value <6). Similar results were found when we investigated the total O:E ratio across different subgroups, using the reported calibration tables and histograms within the included validation studies (appendix 8). Although year of study recruitment and continent did not significantly influence the calibration, we found that miscalibration was more problematic in (developed) countries with low mortality rates (appendix 10). The C statistic did not appear to differ importantly as the standard deviation of the EUROSCORE or age distribution increased.
Fig 5 Results from random effects meta-regression models in the case study (predictive performance of the EuroSCORE for all cause mortality at 30 days in patients undergoing coronary artery bypass grafting). Solid lines=regression lines; dashed lines=95% confidence intervals; dots=included validation studies
Overall, we can conclude that the additive EuroSCORE fairly discriminates between mortality and survival in patients undergoing CABG. Its overall calibration, however, is quite poor because predicted risks appear too high in low risk patients, and the extent of miscalibration substantially varies across populations. Not enough information is available to draw conclusions on the performance of EuroSCORE in high risk patients. Although it has been suggested that overprediction likely occurs due to improvements in cardiac surgery, we could not confirm this effect in the present analyses.
Sensitivity analysis
As for any meta-analysis, it is important to show that results are not distorted by low quality validation studies. For this reason, key analyses should be repeated for the studies at lower and higher risk of bias.
We performed a subgroup analysis by excluding those studies at high risk of bias, to ascertain their effect (fig 2 ⇑ ). Results in table 2 ⇓ indicate that this approach yielded similar summary estimates of discrimination and calibration as those in the full analysis of all studies.
Results from the case study (predictive performance of the EuroSCORE for all cause mortality at 30 days in patients undergoing coronary artery bypass grafting) after excluding studies with high risk of bias
Reporting and presentation
As for any other type of systematic review and meta-analysis, it is important to report the conducted research in sufficient detail. The PRISMA statement (preferred reporting items for systematic reviews and meta-analyses) 41 highlights the key issues for reporting of meta-analysis of intervention studies, which are also generally relevant for meta-analysis of model validation studies. If meta-analysis of individual participant data (IPD) has been used, then PRISMA-IPD will also be helpful. 42 Furthermore, the TRIPOD statement (transparent reporting of a multivariable prediction model for individual prognosis or diagnosis) 23 43 provides several recommendations for the reporting of studies developing, validating, or updating a prediction model, and can be considered here as well. Finally, use of the GRADE approach (grades of recommendation, assessment, development, and evaluation) might help to interpret the results of the systematic review and to present the evidence. 21
As illustrated in this article, researchers should clearly describe the review question, search strategy, tools used for critical appraisal and risk of bias assessment, quality of the included studies, methods used for data extraction and meta-analysis, data used for meta-analysis, and corresponding results and their uncertainty. Furthermore, we recommend to report details on the relevant study populations (eg, using the mean and standard deviation of the linear predictor) and to present summary estimates with confidence intervals and, if appropriate, prediction intervals. Finally, it might be helpful to report probabilities of good performance separately for each performance measure, because researchers can then decide which criteria are most relevant for their situation.
Concluding remarks
In this article, we provide guidance on how to systematically review and quantitatively synthesize the predictive performance of a prediction model. Although we focused on systematic review and meta-analysis of a prognostic model, all guidance can similarly be applied to the meta-analysis of a diagnostic prediction model. We discussed how to define the systematic review question, identify the relevant prediction model studies from the literature, critically appraise the identified studies, extract relevant summary statistics, quantitatively summarise the extracted estimates, and investigate sources of between-study heterogeneity.
Meta-analysis of a prediction model’s predictive performance bears many similarities to other types of meta-analysis. However, in contrast to synthesis of randomised trials, heterogeneity is much more likely in meta-analysis of studies assessing the predictive performance of a prediction model, owing to the increased variation of eligible study designs, increased inclusion of studies with different populations, and increased complexity of required statistical methods. When substantial heterogeneity occurs, summary estimates of model performance can be of limited value. For this reason, it is paramount to identify relevant studies through a systematic review, assess the presence of important subgroups, and evaluate the performance the model is likely to yield in new studies.
Although several concerns can be resolved by aforementioned strategies, it is possible that substantial between-study heterogeneity remains and can only be addressed by harmonising and analysing the study individual participant data. 19 Previous studies have demonstrated that access to individual participant data might also help to retrieve unreported performance measures (eg, calibration slope), estimate the within-study correlation between performance measures, 9 avoid continuity corrections and data transformations, further interpret model generalisability, 8 19 22 31 and tailor the model to populations at hand. 44
Often, multiple models exist for predicting the same condition in similar populations. In such situations, it could be desirable to investigate their relative performance. Although this strategy has already been adopted by several authors, caution is warranted in the absence of individual participant data. In particular, the lack of head-to-head comparisons between competing models and the increased likelihood of heterogeneity across validation studies renders comparative analyses highly prone to bias. Further, it is well known that performance measures such as the C statistic are relatively insensitive to improvements in predictive performance. We therefore believe that summary performance estimates might often be of limited value, and that a meta-analysis should rather focus on assessing their variability across relevant settings and populations. Formal comparisons between competing models are possible (eg, by adopting network meta-analysis methods) but appear most useful for exploratory purposes.
Finally, the following limitations need to be considered in order to fully appreciate this guidance. Firstly, our empirical example demonstrates that the level of reporting in validation studies is often poor. Although the quality of reporting has been steadily improving over the past few years, it will often be necessary to restore missing information from other quantities. This strategy might not always be reliable, such that sensitivity analyses remain paramount in any meta-analysis. Secondly, the statistical methods we discussed in this article are most applicable when meta-analysing the performance results from prediction models developed with logistic regression. Although the same principles apply to survival models, the level of reporting tends to be even less consistent because many more statistical choices and multiple time points need to be considered. Thirdly, we focused on frequentist methods for summarising model performance and calculating corresponding prediction intervals. Bayesian methods have, however, been recommended when predicting the likely performance in a future validation study. 45 Lastly, we mainly focused on statistical measures of model performance, and did not discuss how to meta-analyse clinical measures of performance such as net benefit. 46 Because these performance measures are not frequently reported and typically require subjective thresholds, summarising them appears difficult without access to individual participant data. Nevertheless, further research on how to meta-analyse net benefit estimates would be welcome.
In summary, systematic review and meta-analysis of prediction model performance could help to interpret the potential applicability and generalisability of a prediction model. When the meta-analysis shows promising results, it may be worthwhile to obtain individual participant data to investigate in more detail how the model performs across different populations and subgroups. 19 44
Contributors: KGMM, TPAD, JBR, and RDR conceived the paper objectives. TPAD prepared a first draft of this article, which was subsequently reviewed in multiple rounds by JAAGD, JE, KIES, LH, RDR, JBR, and KGMM. TPAD and JAAGD undertook the data extraction and statistical analyses. TPAD, JAAGD, RDR, and KGMM contributed equally to the paper. All authors approved the final version of the submitted manuscript. TPAD is guarantor. All authors had full access to all of the data (including statistical reports and tables) in the study and can take responsibility for the integrity of the data and the accuracy of the data analysis.
Funding: Financial support received from the Cochrane Methods Innovation Funds Round 2 (MTH001F) and the Netherlands Organization for Scientific Research (91617050 and 91810615). This work was also supported by the UK Medical Research Council Network of Hubs for Trials Methodology Research (MR/L004933/1- R20). RDR was supported by an MRC partnership grant for the PROGnosis RESearch Strategy (PROGRESS) group (grant G0902393). None of the funding sources had a role in the design, conduct, analyses, or reporting of the study or in the decision to submit the manuscript for publication.
Competing interests: All authors have completed the ICMJE uniform disclosure form at www.icmje.org/coi_disclosure.pdf and declare: support from the Cochrane Methods Innovation Funds Round 2, Netherlands Organization for Scientific Research, and the UK Medical Research Council for the submitted work; no financial relationships with any organisations that might have an interest in the submitted work in the previous three years; no other relationships or activities that could appear to have influenced the submitted work.
We thank The BMJ editors and reviewers for their helpful feedback on this manuscript.
- ↵ Khan K, Kunz R, Kleijnen J, et al. Systematic reviews to support evidence-based medicine: how to review and apply findings of healthcare research. CRC Press, 2nd ed, 2011.
- ↵ Steyerberg EW, Moons KGM, van der Windt DA, et al. PROGRESS Group. Prognosis Research Strategy (PROGRESS) 3: prognostic model research. PLoS Med 2013 ; 10 : e1001381 . doi:10.1371/journal.pmed.1001381 pmid:23393430 . OpenUrl CrossRef PubMed
- ↵ Geersing GJ, Bouwmeester W, Zuithoff P, Spijker R, Leeflang M, Moons KG. Search filters for finding prognostic and diagnostic prediction studies in Medline to enhance systematic reviews [correction in: PLoS One 2012;7(7): doi/10.1371/annotation/96bdb520-d704-45f0-a143-43a48552952e]. PLoS One 2012 ; 7 : e32844 . doi:10.1371/journal.pone.0032844 pmid:22393453 . OpenUrl CrossRef PubMed
- ↵ Wong SS, Wilczynski NL, Haynes RB, Ramkissoonsingh R. Hedges Team. Developing optimal search strategies for detecting sound clinical prediction studies in MEDLINE. AMIA Annu Symp Proc 2003 : 728 - 32 . pmid:14728269 .
- ↵ Ingui BJ, Rogers MA. Searching for clinical prediction rules in MEDLINE. J Am Med Inform Assoc 2001 ; 8 : 391 - 7 . doi:10.1136/jamia.2001.0080391 pmid:11418546 . OpenUrl Abstract / FREE Full Text
- ↵ Moons KGM, de Groot JAH, Bouwmeester W, et al. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med 2014 ; 11 : e1001744 . doi:10.1371/journal.pmed.1001744 pmid:25314315 . OpenUrl CrossRef PubMed
- ↵ Wolff R, Whiting P, Mallett S, et al. PROBAST: a risk of bias tool for prediction modelling studies. Cochrane Colloquium Vienna. 2015.
- ↵ Debray TPA, Vergouwe Y, Koffijberg H, Nieboer D, Steyerberg EW, Moons KG. A new framework to enhance the interpretation of external validation studies of clinical prediction models. J Clin Epidemiol 2015 ; 68 : 279 - 89 . doi:10.1016/j.jclinepi.2014.06.018 pmid:25179855 . OpenUrl CrossRef PubMed
- ↵ Snell KI, Hua H, Debray TP, et al. Multivariate meta-analysis of individual participant data helped externally validate the performance and implementation of a prediction model. J Clin Epidemiol 2016 ; 69 : 40 - 50 . doi:10.1016/j.jclinepi.2015.05.009 pmid:26142114 . OpenUrl CrossRef PubMed
- ↵ Altman DG, Royston P. What do we mean by validating a prognostic model? Stat Med 2000 ; 19 : 453 - 73 . doi:10.1002/(SICI)1097-0258(20000229)19:4<453::AID-SIM350>3.0.CO;2-5 pmid:10694730 . OpenUrl CrossRef PubMed Web of Science
- ↵ Justice AC, Covinsky KE, Berlin JA. Assessing the generalizability of prognostic information. Ann Intern Med 1999 ; 130 : 515 - 24 . doi:10.7326/0003-4819-130-6-199903160-00016 pmid:10075620 . OpenUrl CrossRef PubMed Web of Science
- ↵ Collins GS, Omar O, Shanyinde M, Yu LM. A systematic review finds prediction models for chronic kidney disease were poorly reported and often developed using inappropriate methods. J Clin Epidemiol 2013 ; 66 : 268 - 77 . doi:10.1016/j.jclinepi.2012.06.020 pmid:23116690 . OpenUrl CrossRef PubMed
- ↵ Siregar S, Groenwold RHH, de Heer F, Bots ML, van der Graaf Y, van Herwerden LA. Performance of the original EuroSCORE. Eur J Cardiothorac Surg 2012 ; 41 : 746 - 54 . doi:10.1093/ejcts/ezr285 pmid:22290922 . OpenUrl Abstract / FREE Full Text
- ↵ Echouffo-Tcheugui JB, Batty GD, Kivimäki M, Kengne AP. Risk models to predict hypertension: a systematic review. PLoS One 2013 ; 8 : e67370 . doi:10.1371/journal.pone.0067370 pmid:23861760 . OpenUrl CrossRef PubMed
- ↵ Tzoulaki I, Liberopoulos G, Ioannidis JPA. Assessment of claims of improved prediction beyond the Framingham risk score. JAMA 2009 ; 302 : 2345 - 52 . doi:10.1001/jama.2009.1757 pmid:19952321 . OpenUrl CrossRef PubMed Web of Science
- ↵ Eichler K, Puhan MA, Steurer J, Bachmann LM. Prediction of first coronary events with the Framingham score: a systematic review. Am Heart J 2007 ; 153 : 722 - 31, 731.e1-8 . doi:10.1016/j.ahj.2007.02.027 pmid:17452145 . OpenUrl CrossRef PubMed Web of Science
- ↵ Perel P, Edwards P, Wentz R, Roberts I. Systematic review of prognostic models in traumatic brain injury. BMC Med Inform Decis Mak 2006 ; 6 : 38 . doi:10.1186/1472-6947-6-38 pmid:17105661 . OpenUrl CrossRef PubMed
- ↵ Collins GS, de Groot JA, Dutton S, et al. External validation of multivariable prediction models: a systematic review of methodological conduct and reporting. BMC Med Res Methodol 2014 ; 14 : 40 . doi:10.1186/1471-2288-14-40 pmid:24645774 . OpenUrl CrossRef PubMed
- ↵ Riley RD, Ensor J, Snell KI, et al. External validation of clinical prediction models using big datasets from e-health records or IPD meta-analysis: opportunities and challenges. BMJ 2016 ; 353 : i3140 . doi:10.1136/bmj.i3140 pmid:27334381 . OpenUrl FREE Full Text
- ↵ Peat G, Riley RD, Croft P, et al. PROGRESS Group. Improving the transparency of prognosis research: the role of reporting, data sharing, registration, and protocols. PLoS Med 2014 ; 11 : e1001671 . doi:10.1371/journal.pmed.1001671 pmid:25003600 . OpenUrl
- ↵ Iorio A, Spencer FA, Falavigna M, et al. Use of GRADE for assessment of evidence about prognosis: rating confidence in estimates of event rates in broad categories of patients. BMJ 2015 ; 350 : h870 . doi:10.1136/bmj.h870 pmid:25775931 . OpenUrl FREE Full Text
- ↵ Vergouwe Y, Moons KGM, Steyerberg EW. External validity of risk models: Use of benchmark values to disentangle a case-mix effect from incorrect coefficients. Am J Epidemiol 2010 ; 172 : 971 - 80 . doi:10.1093/aje/kwq223 pmid:20807737 . OpenUrl Abstract / FREE Full Text
- ↵ Moons KGM, Altman DG, Reitsma JB, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015 ; 162 : W1-73 . doi:10.7326/M14-0698 pmid:25560730 . OpenUrl PubMed
- ↵ van Klaveren D, Gönen M, Steyerberg EW, Vergouwe Y. A new concordance measure for risk prediction models in external validation settings. Stat Med 2016 ; 35 : 4136 - 52 . doi:10.1002/sim.6997 pmid:27251001 . OpenUrl
- Higgins JPT, Green S. Combining Groups. http://handbook.cochrane.org/chapter_7/7_7_3_8_combining_groups.htm , 2011.
- ↵ Hozo SP, Djulbegovic B, Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol 2005 ; 5 : 13 . doi:10.1186/1471-2288-5-13 pmid:15840177 . OpenUrl CrossRef PubMed
- ↵ Bouwmeester W, Zuithoff NPA, Mallett S, et al. Reporting and methods in clinical prediction research: a systematic review. PLoS Med 2012 ; 9 : 1 - 12 . doi:10.1371/journal.pmed.1001221 pmid:22629234 . OpenUrl PubMed
- ↵ Austin PC, Pencinca MJ, Steyerberg EW. Predictive accuracy of novel risk factors and markers: A simulation study of the sensitivity of different performance measures for the Cox proportional hazards regression model. Stat Methods Med Res 2015 ; 0962280214567141 . pmid:25656552 .
- ↵ Blanche P, Dartigues JF, Jacqmin-Gadda H. Review and comparison of ROC curve estimators for a time-dependent outcome with marker-dependent censoring. Biom J 2013 ; 55 : 687 - 704 . doi:10.1002/bimj.201200045 pmid:23794418 . OpenUrl CrossRef PubMed Web of Science
- ↵ Jinks RC, Royston P, Parmar MKB. Discrimination-based sample size calculations for multivariable prognostic models for time-to-event data. BMC Med Res Methodol 2015 ; 15 : 82 . doi:10.1186/s12874-015-0078-y pmid:26459415 . OpenUrl CrossRef PubMed
- ↵ Pennells L, Kaptoge S, White IR, Thompson SG, Wood AM. Emerging Risk Factors Collaboration. Assessing risk prediction models using individual participant data from multiple studies. Am J Epidemiol 2014 ; 179 : 621 - 32 . doi:10.1093/aje/kwt298 pmid:24366051 . OpenUrl Abstract / FREE Full Text
- ↵ Riley RD, Higgins JPT, Deeks JJ. Interpretation of random effects meta-analyses. BMJ 2011 ; 342 : d549 . doi:10.1136/bmj.d549 pmid:21310794 . OpenUrl FREE Full Text
- ↵ Snell KIE. Development and application of statistical methods for prognosis research. PhD thesis, School of Health and Population Sciences, Birmingham, UK, 2015.
- ↵ van Klaveren D, Steyerberg EW, Perel P, Vergouwe Y. Assessing discriminative ability of risk models in clustered data. BMC Med Res Methodol 2014 ; 14 : 5 . doi:10.1186/1471-2288-14-5 pmid:24423445 . OpenUrl CrossRef PubMed
- ↵ Gengsheng Qin , Hotilovac L. Comparison of non-parametric confidence intervals for the area under the ROC curve of a continuous-scale diagnostic test. Stat Methods Med Res 2008 ; 17 : 207 - 21 . doi:10.1177/0962280207087173 pmid:18426855 . OpenUrl Abstract / FREE Full Text
- ↵ IntHout J, Ioannidis JP, Borm GF. The Hartung-Knapp-Sidik-Jonkman method for random effects meta-analysis is straightforward and considerably outperforms the standard DerSimonian-Laird method. BMC Med Res Methodol 2014 ; 14 : 25 . doi:10.1186/1471-2288-14-25 pmid:24548571 . OpenUrl CrossRef PubMed
- ↵ Cornell JE, Mulrow CD, Localio R, et al. Random-effects meta-analysis of inconsistent effects: a time for change. Ann Intern Med 2014 ; 160 : 267 - 70 . doi:10.7326/M13-2886 pmid:24727843 . OpenUrl PubMed Web of Science
- ↵ Ban JW, Emparanza JI, Urreta I, Burls A. Design Characteristics Influence Performance of Clinical Prediction Rules in Validation: A Meta-Epidemiological Study. PLoS One 2016 ; 11 : e0145779 . doi:10.1371/journal.pone.0145779 pmid:26730980 . OpenUrl
- ↵ Deeks JJ, Higgins JPT, Altman DG. Chapter 9. Analysing data and undertaking meta-analyses. Cochrane Collaboration, 2011 .
- ↵ Berlin JA, Santanna J, Schmid CH, Szczech LA, Feldman HI. Anti-Lymphocyte Antibody Induction Therapy Study Group. Individual patient- versus group-level data meta-regressions for the investigation of treatment effect modifiers: ecological bias rears its ugly head. Stat Med 2002 ; 21 : 371 - 87 . doi:10.1002/sim.1023 pmid:11813224 . OpenUrl CrossRef PubMed Web of Science
- ↵ Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 2009 ; 339 : b2700 . doi:10.1136/bmj.b2700 pmid:19622552 . OpenUrl Abstract / FREE Full Text
- ↵ Stewart LA, Clarke M, Rovers M, et al. PRISMA-IPD Development Group. Preferred reporting items for a systematic review and meta- analysis of individual participant data: The PRISMA-IPD Statement. JAMA 2015 ; 313 : 1657 - 65 . doi:10.1001/jama.2015.3656 pmid:25919529 . OpenUrl CrossRef PubMed
- ↵ Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med 2015 ; 162 : 55 - 63 . doi:10.7326/M14-0697 pmid:25560714 . OpenUrl CrossRef PubMed
- ↵ Debray TPA, Riley RD, Rovers MM, Reitsma JB, Moons KG. Cochrane IPD Meta-analysis Methods group. Individual participant data (IPD) meta-analyses of diagnostic and prognostic modeling studies: guidance on their use. PLoS Med 2015 ; 12 : e1001886 . doi:10.1371/journal.pmed.1001886 pmid:26461078 . OpenUrl CrossRef PubMed
- ↵ Sutton AJ, Abrams KR. Bayesian methods in meta-analysis and evidence synthesis. Stat Methods Med Res 2001 ; 10 : 277 - 303 . doi:10.1191/096228001678227794 pmid:11491414 . OpenUrl Abstract / FREE Full Text
- ↵ Vickers AJ, Van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ 2016 ; 352 : i6 . doi:10.1136/bmj.i6 pmid:26810254 . OpenUrl FREE Full Text
- AI Collection
- Oxford Thesis Collection
- CC0 version of this metadata
Deep learning for time series prediction and decision making over time
In this thesis, we develop a collection of state-of-the-art deep learning models for time series forecasting. Primarily focusing on a closer alignment with traditional methods in time series modelling, we adopt three main directions of research -- 1) novel architectures, 2) hybrid models, and 3) feature extraction. Firstly, we propose two new architectures for general one-step-ahead and multi-horizon forecasting. With the Recurrent Neural Filter (RNF), we take a closer look at the relation...
Email this record
Please enter the email address that the record information will be sent to.
Please add any additional information to be included within the email.
Cite this record
Chicago style, access document.
- DPhil_Thesis.pdf (Dissemination version, Version of record, 14.5MB)
Why is the content I wish to access not available via ORA?
Content may be unavailable for the following four reasons.
- Version unsuitable We have not obtained a suitable full-text for a given research output. See the versions advice for more information.
- Recently completed Sometimes content is held in ORA but is unavailable for a fixed period of time to comply with the policies and wishes of rights holders.
- Permissions All content made available in ORA should comply with relevant rights, such as copyright. See the copyright guide for more information.
- Clearance Some thesis volumes scanned as part of the digitisation scheme funded by Dr Leonard Polonsky are currently unavailable due to sensitive material or uncleared third-party copyright content. We are attempting to contact authors whose theses are affected.
Alternative access to the full-text
Request a copy.
We require your email address in order to let you know the outcome of your request.
Provide a statement outlining the basis of your request for the information of the author.
Please note any files released to you as part of your request are subject to the terms and conditions of use for the Oxford University Research Archive unless explicitly stated otherwise by the author.
Contributors
Bibliographic details, item description, related items, terms of use, views and downloads.
If you are the owner of this record, you can report an update to it here: Report update to this record
Report an update
We require your email address in order to let you know the outcome of your enquiry.
Review of Water Quality Prediction Methods
- Conference paper
- First Online: 31 May 2023
- Cite this conference paper

- Zhen Chen 9 , 10 ,
- Limin Liu 9 , 10 ,
- Yongsheng Wang 9 , 10 , 10 &
- Jing Gao 11
Part of the book series: Lecture Notes in Civil Engineering ((LNCE,volume 341))
Included in the following conference series:
- International Conference on Water Resource and Environment
222 Accesses
1 Citations
Water quality prediction plays a crucial role in environmental monitoring, ecosystem sustainability, and aquaculture, and it plays an important role in both economic and ecological benefits. For many years, researchers have been working on how to improve the accuracy of water quality prediction. However, at this stage, with the increase in external climate change, external noise, precipitation, and many other uncertainties, water quality prediction is facing the problem of insufficient accuracy. This paper analyzes the research methods of water quality prediction at home and abroad in recent years and summarizes the introduction through two aspects: mechanical water quality prediction methods and non-mechanical water quality prediction methods. Firstly, the mechanism of water quality prediction method is introduced, which uses various hydrological data information such as initial water head, bottom slope, hydraulic radius, etc. to predict water quality; Secondly, it introduces the non-mechanical water quality prediction method that uses historical water quality index data to analyze and mine to predict water quality. Finally, after introducing the existing prediction methods, the development direction of water quality prediction is analyzed and summarized.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
- Available as PDF
- Read on any device
- Instant download
- Own it forever
- Available as EPUB and PDF
- Durable hardcover edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Han HG, Qiao JF, Chen QL (2012) Model predictive control of dissolved oxygen concentration based on a self-organizing RBF neural network. Control Eng Pract 20(4):465–476
Google Scholar
Wang Y, Zhou J, Chen K, Wang Y, Liu L (2017) Water quality prediction method based on LSTM neural network. In: 2017 12th international conference on intelligent systems and knowledge engineering (ISKE). IEEE, pp 1–5
Wang XY (2015) Research on urban water supply quality monitoring and early warning system
Fan M, Gu ZL (2010) Research progress and development trend of water quality model. Shanghai Environ Sci
Rinaldi S, Soncini-Sessa R (1978) Sensitivity analysis of generalized Streeter-Phelps models. Adv Water Resour 1(3):141–146
Xiao YJ (2018) The suitability of WASP model and QUAL2K model to simulate the water quality of Beichuan River
Miao HY, Zhou XC (2006) Application of improved genetic algorithm in parameter calibration of S-P BOD-DO water quality model. Chin J Hydraul Archit Eng (01):67–69
He Q, Sun SQ, Nie L (2003) Estimation of parameters K_1 and K_2 of S-P model for water quality of Nanfei River. J Hefei Univ Technol (Nat Sci Edn) 02:286–290
Liu BX, Lai XL, Cao Q, Cheng X (2013) In advanced materials research, vol 610, pp 1705–1709
Li SL, Li GY (2014) Research on the establishment of water resources management model coupled with S-P water quality simulation under uncertain conditions. Northeast Water Resour Hydropower (06):33–35+72
Wu J, Yu X (2021) Numerical investigation of dissolved oxygen transportation through a coupled SWE and Streeter–Phelps model
Paliwal R, Sharma P, Kansal A (2007) Water quality modelling of the river Yamuna (India) using QUAL2E-UNCAS. J Environ Manage 83(2):131–144
CAS PubMed Google Scholar
Bárbara VF (2006) Uso do modelo QUAL2E no estudo da qualidade da água e da capacidade de autodepuração do Rio Araguari-AP (Amazônia)
Knapik H, Fernandes C, Pickbrenner K, Porto M, Bassanesi K (2011) Qualidade da água da bacia do rio Iguaçu: diferenças conceituais entre os modelos QUAL2E e QUAL2K. Revista Brasileira de Recursos Hídricos 16(2):75–88
Zhao YX, Chen Y, Wu YY (2015) Theoretical method and application guide of QUAL2K river water quality simulation model. Meteorological Press
Gong QL (2016) Uncertainty study on the parameters of QUAL2K water quality model
Chen QS, Xie XH, Du QY, Liu Y (2018) Parameters sensitivity analysis of DO in water quality model of QUAL2K. In: IOP Conference series: earth and environmental science, vol 191, no 1. IOP Publishing, p 012030
Kuczera G, Diment G (1988) General water supply system simulation model: WASP. J Water Resour Plan Manag 114(4):365–382
Ambrose RB, Wool TA, Martin JL (1993) The water quality analysis simulation program, WASP5, Part A: model documentation. Environ Research Laboratory, US Environmental Protection Agency, Athens, GA
Dai TJ (2019) Research on water quality evaluation and prediction of Taolinkou reservoir
Liu C (2020) Evaluation of the purification capacity of typical Baiyangdian waters based on WASP water quality prediction model
Zhao ZH, Yao J (2021) Research on water pollution control of mountain rivers based on WASP model. People’s Yangtze River S1:38–41
Wang W, Wang X (2020) Progress in classification and application of coupled water quality models. People's Pearl River (07):79–84
He Q, Li JJ, Huang L (2014) Scenario analysis of water quality in the main section of the Yangtze River-Jialing River in Chongqing based on the coupled model of EFDC and WASP. In: 2014 annual meeting of the chinese society for environmental sciences (Chapter 4), pp 888–895
Jia H, Wang S, Wei M, Zhang Y (2011) Scenario analysis of water pollution control in the typical peri-urban river using a coupled hydrodynamic-water quality model. Front Environ Sci Eng China 5(2):255–265
CAS Google Scholar
Douglas-Mankin KR, Srinivasan R, Arnold JG (2010) Soil and Water Assessment Tool (SWAT) model: current developments and applications. Trans ASABE 53(5):1423–1431
de Andrade CW, Montenegro SM, Montenegro AA, Lima JRDS, Srinivasan R, Jones CA (2019) Soil moisture and discharge modeling in a representative watershed in northeastern Brazil using SWAT. Ecohydrol Hydrobiol 19(2):238–251
Pradhan P, Tingsanchali T, Shrestha S (2020) Evaluation of soil and water assessment tool and artificial neural network models for hydrologic simulation in different climatic regions of Asia. Sci Total Environ 701:134308
Giles NA, Babbar-Sebens M, Srinivasan R, Ficklin DL, Barnhart B (2019) Optimization of linear stream temperature model parameters in the soil and water assessment tool for the continental United States. Ecol Eng 127:125–134
Kalcic MM, Chaubey I, Frankenberger J (2015) Defining Soil and Water Assessment Tool (SWAT) hydrologic response units (HRUs) by field boundaries. Int J Agric Biol Eng 8(3):69–80
Li J, Ma TX, Lu YR, Song XF, Li RK, Liu JZ, Duan Z (2021) Multi-objective calibration and evaluation of SWAT model: Taking Meichuan River Basin as an example. J Univ Chin Acad Sci 05:590–600
Zhang ZM, Wang XY, Pan R (2017) An improved method for parameter calibration of uncertain water quality models. China Environ Sci (03):956–962
Shaw AR, Smith Sawyer H, LeBoeuf EJ, McDonald MP, Hadjerioua B (2017) Hydropower optimization using artificial neural network surrogate models of a high-fidelity hydrodynamics and water quality model. Water Resour Res 53(11):9444–9461
Xiang SL, Liu ZM, You BS (2006) Study on multiple linear regression analysis model for groundwater flow prediction. Hydrology 06:36–37
Chen ZM, Wang W, Zhao Y, Xu ZY (2020) Improved principal component analysis and multiple regression fusion for Hanfeng Lake water quality assessment and prediction. Environ Monitor Manage Technol 32(4):5
Teng EJ, Liu TL, An H (1995) Compilation of conversion method for non-ionic ammonia of surface water environmental quality standard. China Environ Monitor (04)
Nourani V, Alami MT, Vousoughi FD (2016) Self-organizing map clustering technique for ANN-based spatiotemporal modeling of groundwater quality parameters. J Hydroinf 18(2):288–309
Liu TQ, Wang QL (2017) Bayesian prediction of DO and NH 4 + -N content in Xiangjiang River Basin based on ARIMA model. J Hunan Agric Univ (Natural Science Edition) 05:575–580
Melesse AM, Ahmad S, McClain ME, Wang X, Lim YH (2011) Suspended sediment load prediction of river systems: an artificial neural network approach. Agric Water Manag 98(5):855–866
Zare A, Bayat V, Daneshkare A (2011) Forecasting nitrate concentration in groundwater using artificial neural network and linear regression models. Int Agrophys 25(2)
Huo S, He Z, Su J, Xi B, Zhu C (2013) Using artificial neural network models for eutrophication prediction. Procedia Environ Sci 18:310–316
Chang FJ, Chen PA, Chang LC, Tsai YH (2016) Estimating spatio-temporal dynamics of stream total phosphate concentration by soft computing techniques. Sci Total Environ 562:228–236
Rajaee T, Khani S, Ravansalar M (2020) Artificial intelligence-based single and hybrid models for prediction of water quality in rivers: a review. Chemom Intell Lab Syst 200:103978
Wang LX (2021) Research on anomaly detection method of urban river water quality based on multi-index time series data
Liu LX, Sun QX, Wang SY (1989) A preliminary study on the application of grey system theory to comprehensive evaluation of new crop varieties. Chin Agric Sci (03):22–27
Zhang DY (2021) Research and application of water quality prediction method based on EEMD-LSTM
Hu ZB, Pang Y, Song WW, Shao YX (2019) Application of grey system dynamic model group GM (1,1) in water quality prediction of Qinhuai River. Sichuan Environ 01:116–119
Delgado A, Vriclizar D, Medina E (2017) Artificial intelligence model based on grey systems to assess water quality from Santa River watershed. In: 2017 electronic congress (E-CON UNI). IEEE, pp 1–4
Li S, Zeng B, Ma X, Zhang D (2020) A novel grey model with a three-parameter background value and its application in forecasting average annual water consumption per capita in urban areas along the Yangtze River Basin. J Grey Syst 32(1)
Zuo K (2021) A new generalized discrete grey prediction model and its application. Math Pract Understand 24:1–13
Haghiabi AH, Nasrolahi AH, Parsaie A (2018) Water quality prediction using machine learning methods. Water Qual Res J 53(1):3–13
Khani S, Rajaee T (2017) Modeling of dissolved oxygen concentration and its hysteresis behavior in rivers using wavelet transform‐based hybrid models. CLEAN–Soil, Air, Water 45(2)
Sun T, Pan SB, Li YJ (2004) Application of artificial neural network model in evaluation and classification of groundwater quality. Hydrogeol Eng Geol 03:58–61
Zhao Y, Nan J, Cui FY, Guo L (2007) Water quality forecast through application of BP neural network at Yuqiao reservoir. J Zhejiang Univ–Sci A 8(9):1482–1487
Ahmed AN, Othman FB, Afan HA, Ibrahim RK, Fai CM, Hossain MS, Elshafie A (2019) Machine learning methods for better water quality prediction. J Hydrol 578:124084
Karsoliya S (2012) Approximating number of hidden layer neurons in multiple hidden layer BPNN architecture. Int J Eng Trends Technol 3(6):714–717
Yu YJ, Jiang WG, Xu MF (2011) Prediction of chlorophyll a in water by BP neural network based on PSO algorithm. Environ Sci Res 05:526–532
Ding YR, Cai YJ, Sun PD, Chen B (2014) The use of combined neural networks and genetic algorithms for prediction of river water quality. J Appl Res Technol 12(3):493–499
Yan J, Xu Z, Yu Y, Xu H, Gao K (2019) Application of a hybrid optimized BP network model to estimate water quality parameters of Beihai Lake in Beijing. Appl Sci 9(9):1863
Deng Y, Zhou X, Shen J, Xiao G, Hong H, Lin H, Liao BQ (2021) New methods based on back propagation (BP) and radial basis function (RBF) artificial neural networks (ANNs) for predicting the occurrence of haloketones in tap water. Sci Total Environ 772:145534
Yaseen ZM, El-Shafie A, Afan HA, Hameed M, Mohtar WHMW, Hussain A (2016) RBFNN versus FFNN for daily river flow forecasting at Johor River, Malaysia. Neural Comput Appl 27(6):1533–1542
Lin H, Dai Q, Zheng L, Hong H, Deng W, Wu F (2020) Radial basis function artificial neural network able to accurately predict disinfection by-product levels in tap water: taking haloacetic acids as a case study. Chemosphere 248:125999
Meng X, Zhang Y, Qiao J (2021) An adaptive task-oriented RBF network for key water quality parameters prediction in wastewater treatment process. Neural Comput Appl 33(17):11401–11414
Yang H, Wang X, Sun J, Li D (2020) Dissolved oxygen prediction using RBF network based on improved conjugate gradient method. In: 2020 IEEE 11th international conference on software engineering and service science (ICSESS). IEEE, pp 515–518
Ren Z, Li W, Qiao J (2018) A recurrent RBF neural network based on modified gravitational search algorithm. In: 2018 Chinese Automation Congress (CAC). IEEE, pp 4079–4083
Ghose DK, Panda SS, Swain PC (2010) Prediction of water table depth in western region, Orissa using BPNN and RBFN neural networks. J Hydrol 394(3–4):296–304
Suen JP, Eheart JW (2003) Evaluation of neural networks for modeling nitrate concentrations in rivers. J Water Resour Plan Manag 129(6):505–510
Hong H, Zhang Z, Guo A, Shen L, Sun H, Liang Y, Lin H (2020) Radial basis function artificial neural network (RBF ANN) as well as the hybrid method of RBF ANN and grey relational analysis able to well predict trihalomethanes levels in tap water. J Hydrol 591:125574
Wu D, Wang H, Seidu R (2020) Smart data driven quality prediction for urban water source management. Futur Gener Comput Syst 107:418–432
Chen L, Hu X, Xu T, Kuang H, Li Q (2017) Turn signal detection during nighttime by CNN detector and perceptual hashing tracking. IEEE Trans Intell Transp Syst 18(12):3303–3314
Jia WS, Zhang HZ, Jie MA, Liang G, Wang JH, Xin LIU (2020) Study on the predication modeling of COD for water based on UV-VIS Spectroscopy and CNN Algorithm of Deep Learning. Spectrosc Spectral Anal 40(9):2981
Pyo J, Park LJ, Pachepsky Y, Baek SS, Kim K, Cho KH (2020) Using convolutional neural network for predicting cyanobacteria concentrations in river water. Water Res 186:116349
Wang L, Wu Y, Xu J, Zhang H, Wang X, Yu J, Zhao Z (2020) Status prediction by 3d fractal net CNN based on remote sensing images. Fractals 28(08):2040018
Song CM, Kim JS (2020) Applicability evaluation of the hydrological image and convolution neural network for prediction of the biochemical oxygen demand and total phosphorus loads in agricultural areas. Agric 10(11):529
Lim H, An H, Kim H, Lee J (2019) Prediction of pollution loads in the Geum River upstream using the recurrent neural network algorithm. Korean J Agric Sci 46(1):67–78
Hu Z, Zhang Y, Zhao Y, Xie M, Zhong J, Tu Z, Liu J (2019) A water quality prediction method based on the deep LSTM network considering correlation in smart mariculture. Sensors 19(6):1420
PubMed PubMed Central Google Scholar
Cao X, Liu Y, Wang J, Liu C, Duan Q (2020) Prediction of dissolved oxygen in pond culture water based on K-means clustering and gated recurrent unit neural network. Aquacult Eng 91:102122
Hu YK, Wang N, Liu S, Jiang QL, Zhang N (2021) Application of time series model and LSTM model in water quality prediction. Small Microcomput Syst 08:1569–1573
Eze E, Ajmal T (2020) Dissolved oxygen forecasting in aquaculture: a hybrid model approach. Appl Sci 10(20):7079
Yang Y, Xiong Q, Wu C, Zou Q, Yu Y, Yi H, Gao M (2021) A study on water quality prediction by a hybrid CNN-LSTM model with attention mechanism. Environ Sci Pollut Res 28(39):55129–55139
Zhou CM, Liu MP, Wang JW (2021) Water quality prediction model based on CNN-LSTM. Hydropower Energy Sci 39(03):20–23
Zhou J, Wang Y, Xiao F, Wang Y, Sun L (2018) Water quality prediction method based on IGRA and LSTM. Water 10(9):1148
Li Z, Peng F, Niu B, Li G, Wu J, Miao Z (2018) Water quality prediction model combining sparse auto-encoder and LSTM network. IFAC-PapersOnLine 51(17):831–836
Singh KP, Basant A, Malik A, Jain G (2009) Artificial neural network modeling of the river water quality—a case study. Ecol Model 220(6):888–895
Goh AT (1995) Back-propagation neural networks for modeling complex systems. Artif Intell Eng 9(3):143–151
Yang X, Zhang H, Zhou H (2014) A hybrid methodology for salinity time series forecasting based on wavelet transform and NARX neural networks. Arab J Sci Eng 39(10):6895–6905
Qian Z, Pei Y, Cao LX, Wang JY, Jing BO (2016) Overview of wind power prediction methods. High Voltage Technol (04):1047–1060
Parmar KS, Bhardwaj R (2014) Water quality management using statistical analysis and time-series prediction model. Appl Water Sci 4(4):425–434
Download references
Author information
Authors and affiliations.
College of Data Science and Application, Inner Mongolia University of Technology, Hohhot, China
Zhen Chen, Limin Liu & Yongsheng Wang
Inner Mongolia Autonomous Region Software Service Engineering Technology Research Center Based on Big Data Inner Mongolia Autonomous Region, Hohhot, China
Zhen Chen, Limin Liu, Yongsheng Wang & Yongsheng Wang
School of Computer and Information Engineering, Inner Mongolia Agricultural University, Hohhot, China
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Yongsheng Wang .
Editor information
Editors and affiliations.
Department of Civil Engineering, I-Shou University, Kaohsiung, Taiwan
Chih-Huang Weng
Rights and permissions
Reprints and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper.
Chen, Z., Liu, L., Wang, Y., Gao, J. (2023). Review of Water Quality Prediction Methods. In: Weng, CH. (eds) Proceedings of the 8th International Conference on Water Resource and Environment. WRE 2022. Lecture Notes in Civil Engineering, vol 341. Springer, Singapore. https://doi.org/10.1007/978-981-99-1919-2_17
Download citation
DOI : https://doi.org/10.1007/978-981-99-1919-2_17
Published : 31 May 2023
Publisher Name : Springer, Singapore
Print ISBN : 978-981-99-1918-5
Online ISBN : 978-981-99-1919-2
eBook Packages : Engineering Engineering (R0)
Share this paper
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
- Link to facebook
- Link to linkedin
- Link to twitter
- Link to youtube
- Writing Tips
How to Justify Your Methods in a Thesis or Dissertation

4-minute read
- 1st May 2023
Writing a thesis or dissertation is hard work. You’ve devoted countless hours to your research, and you want your results to be taken seriously. But how does your professor or evaluating committee know that they can trust your results? You convince them by justifying your research methods.
What Does Justifying Your Methods Mean?
In simple terms, your methods are the tools you use to obtain your data, and the justification (which is also called the methodology ) is the analysis of those tools. In your justification, your goal is to demonstrate that your research is both rigorously conducted and replicable so your audience recognizes that your results are legitimate.
The formatting and structure of your justification will depend on your field of study and your institution’s requirements, but below, we’ve provided questions to ask yourself as you outline your justification.
Why Did You Choose Your Method of Gathering Data?
Does your study rely on quantitative data, qualitative data, or both? Certain types of data work better for certain studies. How did you choose to gather that data? Evaluate your approach to collecting data in light of your research question. Did you consider any alternative approaches? If so, why did you decide not to use them? Highlight the pros and cons of various possible methods if necessary. Research results aren’t valid unless the data are valid, so you have to convince your reader that they are.
How Did You Evaluate Your Data?
Collecting your data was only the first part of your study. Once you had them, how did you use them? Do your results involve cross-referencing? If so, how was this accomplished? Which statistical analyses did you run, and why did you choose them? Are they common in your field? How did you make sure your data were statistically significant ? Is your effect size small, medium, or large? Numbers don’t always lend themselves to an obvious outcome. Here, you want to provide a clear link between the Methods and Results sections of your paper.
Did You Use Any Unconventional Approaches in Your Study?
Most fields have standard approaches to the research they use, but these approaches don’t work for every project. Did you use methods that other fields normally use, or did you need to come up with a different way of obtaining your data? Your reader will look at unconventional approaches with a more critical eye. Acknowledge the limitations of your method, but explain why the strengths of the method outweigh those limitations.
Find this useful?
Subscribe to our newsletter and get writing tips from our editors straight to your inbox.
What Relevant Sources Can You Cite?
You can strengthen your justification by referencing existing research in your field. Citing these references can demonstrate that you’ve followed established practices for your type of research. Or you can discuss how you decided on your approach by evaluating other studies. Highlight the use of established techniques, tools, and measurements in your study. If you used an unconventional approach, justify it by providing evidence of a gap in the existing literature.
Two Final Tips:
● When you’re writing your justification, write for your audience. Your purpose here is to provide more than a technical list of details and procedures. This section should focus more on the why and less on the how .
● Consider your methodology as you’re conducting your research. Take thorough notes as you work to make sure you capture all the necessary details correctly. Eliminating any possible confusion or ambiguity will go a long way toward helping your justification.
In Conclusion:
Your goal in writing your justification is to explain not only the decisions you made but also the reasoning behind those decisions. It should be overwhelmingly clear to your audience that your study used the best possible methods to answer your research question. Properly justifying your methods will let your audience know that your research was effective and its results are valid.
Want more writing tips? Check out Proofed’s Writing Tips and Academic Writing Tips blogs. And once you’ve written your thesis or dissertation, consider sending it to us. Our editors will be happy to check your grammar, spelling, and punctuation to make sure your document is the best it can be. Check out our services for free .
Share this article:
Post A New Comment
Got content that needs a quick turnaround? Let us polish your work. Explore our editorial business services.
9-minute read
How to Use Infographics to Boost Your Presentation
Is your content getting noticed? Capturing and maintaining an audience’s attention is a challenge when...
8-minute read
Why Interactive PDFs Are Better for Engagement
Are you looking to enhance engagement and captivate your audience through your professional documents? Interactive...
7-minute read
Seven Key Strategies for Voice Search Optimization
Voice search optimization is rapidly shaping the digital landscape, requiring content professionals to adapt their...
Five Creative Ways to Showcase Your Digital Portfolio
Are you a creative freelancer looking to make a lasting impression on potential clients or...
How to Ace Slack Messaging for Contractors and Freelancers
Effective professional communication is an important skill for contractors and freelancers navigating remote work environments....
3-minute read
How to Insert a Text Box in a Google Doc
Google Docs is a powerful collaborative tool, and mastering its features can significantly enhance your...

Make sure your writing is the best it can be with our expert English proofreading and editing.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 25 July 2022
A prediction-focused approach to personality modeling
- Gal Lavi 1 ,
- Jonathan Rosenblatt 2 &
- Michael Gilead 3
Scientific Reports volume 12 , Article number: 12650 ( 2022 ) Cite this article
3160 Accesses
6 Altmetric
Metrics details
- Human behaviour
- Social neuroscience
In the current study, we set out to examine the viability of a novel approach to modeling human personality. Research in psychology suggests that people’s personalities can be effectively described using five broad dimensions (the Five-Factor Model; FFM); however, the FFM potentially leaves room for improved predictive accuracy. We propose a novel approach to modeling human personality that is based on the maximization of the model’s predictive accuracy. Unlike the FFM, which performs unsupervised dimensionality reduction, we utilized a supervised machine learning technique for dimensionality reduction of questionnaire data, using numerous psychologically meaningful outcomes as data labels (e.g., intelligence, well-being, sociability). The results showed that our five-dimensional personality summary, which we term the “Predictive Five” (PF), provides predictive performance that is better than the FFM on two independent validation datasets, and on a new set of outcome variables selected by an independent group of psychologists. The approach described herein has the promise of eventually providing an interpretable, low-dimensional personality representation, which is also highly predictive of behavior.
Similar content being viewed by others

Dimensionality reduction beyond neural subspaces with slice tensor component analysis
Sleep quality, duration, and consistency are associated with better academic performance in college students
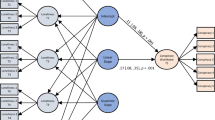
Loneliness trajectories over three decades are associated with conspiracist worldviews in midlife
Introduction.
Humans significantly differ from each other. Some people’s idea of fun is partying all night long, and others enjoy binging on a TV series while eating snacks; some are extremely intelligent, and others less so; some are hot-headed, and others remain cool, no matter what. Because of this variety, predicting humans’ thoughts, feelings, and behaviors is a cumbersome task; nonetheless, we attempt to solve this task on a daily basis. For example, when we decide who to marry, we try to predict whether we can depend on the other person till death do us part; when we choose a career, we must do our best to predict whether we will be successful and fulfilled in a given profession.
In order to predict a person’s thoughts, feelings, and behaviors, people often have no other option but to generate something akin to a scientific theory 1 —a parsimonious model that attempts to capture the unique characteristics of individuals, and that could be used to predict their behavior in novel circumstances. Indeed, research shows that people employ such theories when predicting their own 2 and others’ behaviors. Unfortunately, theories based strictly on intuition are often highly inaccurate 3 , even if produced by professional psychological theoreticians 4 . In light of this, ever since the early days of psychology research, scholars have been attempting to devise personality models using the scientific method, giving rise to the longstanding field of personality science.
Personality, when used as a scientific term, refers to the mental features of individuals that characterize them across different situations, and thus can be used to predict their behavior. In the early years of personality research, scientists generated numerous competing theories and measures, but struggled to arrive at a scientific consensus regarding the core structure of human personality. In recent decades, a consensus theory of the core dimensions of human personality has emerged—the Five Factor Model (FFM).
The FMM emerged from the so-called “lexical paradigm”, which assumed that if people regularly exhibit a form of behavior that is meaningful to human life, then language will produce a term to describe it 5 . Given this assumption, personality psychologists performed research wherein they asked individuals to rate themselves on lists of common English language trait words (e.g., friendly, upbeat), and then developed and used early dimensionality-reduction methods to find a parsimonious model that can account for much of the variability in each person’s trait ratings 5 .
Much research shows that these five factors, often termed the “Big Five” are relatively stable over time and have convergent and discriminant validity across methods and observers 6 . Moreover, research into the FFM has replicated the dimensional structure in different samples, languages, and cultures 7 , 8 (but see 9 for a recent criticism). In light of this, the FFM is taken by some to reflect a comprehensive ontology of the psychological makeup of human beings 10 according to Mccrae and Costa 11 the five factors are “both necessary and reasonably sufficient for describing at a global level the major features of personality’’.
Surely, human beings are complex entities, and their personality is not fully captured by five dimensions; however, the importance of having a parsimonious model of humans’ psychological diversity cannot be overstated. As noted by John and Srivasta 12 , a parsimonious taxonomy permits researchers to study “ specified domains of personality characteristics, rather than examining separately the thousands of particular attributes that make human beings individual and unique.” Moreover, as they note, such a taxonomy greatly facilitate s “ the accumulation and communication of empirical findings by offering a standard vocabulary, or nomenclature”.
An additional consequence of having a parsimonious model of the core dimensions of human personality, is that such an abstraction enables the acquisition of novel knowledge via statistical learning (see 13 for a discussion of the importance of abstract representations in learning); namely, whereas the estimation of covariances between high-dimensional vectors is often highly unreliable (i.e., the so-called “curse of dimensionality” 14 ), learning the statistical correlates of a low-dimensional structure is a more tractable problem. For example, research has shown that participants’ self-reported ratings on the FFM dimensions can be reliably estimated based on their digital footprint 15 .
This ability to infer individuals’ personality traits using machine learning also raises serious concerns, as it may be used for effective psychological manipulation of the public. In 2013, a private company named Cambridge Analytica harvested the data of Facebook users, and used statistical methods to infer the personality characteristics of hundreds of millions of Americans 16 . This psychological profile of the American population was supposedly used by the Trump campaign in an attempt to tailor political advertisements based on an individuals’ specific personality profile. While the success of these methods remains unclear, given the vast amount of data accumulated by companies such as Alphabet and Meta, the potential dangers of machine-learning based psychological profiling is taken by many to be a serious threat to democracy 17 .
Even if dubious entities indeed manage to acquire the Big Five personality profile of entire populations, it is far from obvious that such information could be used to generate actionable predictions. Indeed, the FFM was criticized by some researchers for its somewhat limited contribution to predicting outcomes on meaningful dimensions 18 , 19 , 20 . In light of such claims, some have argued that the public concern over the Cambridge Analytica scandal was overblown 21 (but see 22 for evidence for potential reasons for concern).
Roberts et al. 23 present counter-argument for critical stances against the predictive accuracy of the FFM and note that: “As research on the relative magnitude of effects has documented, personality psychologists should not apologize for correlations between 0.10 and 0.30, given that the effect sizes found in personality psychology are no different than those found in other fields of inquiry.” While this claim is clearly true, there is also no doubt that such correlations (that translate to explained variance in the range of 1%-9%) potentially leave room for improvements in terms of predictive accuracy.
If one’s goal is to find a parsimonious representation of personality that has better predictive accuracy than the FFM, it could be instructive to remember that the statistical method by which the FFM was produced—namely, Factor Analysis—is not geared towards prediction. Factor analysis is an unsupervised dimensionality-reduction method (i.e., a method that maps original data to a new lower dimensional space without utilizing information regarding outcomes) aimed at maximizing explanatory coherence and semantic interpretability, rather than maximizing predictive ability. It does so by finding a parsimonious, low-dimension representation (e.g., the five Big Five factors: extraversion, neuroticism and so on) that maximizes the variance explained in the higher-dimension domain (e.g., hundreds of responses to questionnaire items; for example, “I am lazy”; “I enjoy meeting new people”). Advances in statistics and machine learning have opened up new techniques for supervised dimensionality-reduction. Namely, methods that reduce the dimensionality of a source domain (i.e., predictor variables, \({X}_{1},...{,X}_{n}\) ; in the case of personality, hundreds of questionnaire items) by focusing on the objective of maximizing the capacity of the lower-dimensional representation to predict outcomes of a target domain (outcome variables, \({Y}_{1},...{,Y}_{m}\) , for example, depression, risky behavior, workplace performance).
Such techniques where dimensionality-reduction is achieved via maximization of predictive accuracy across a host of target-domain outcomes hold the potential of providing psychologists with parsimonious models of a psychological feature space that serve as relatively “generalizable predictors” of important aspects of human behavior. Moreover, it may demonstrate that privacy leaks, a-lá Cambridge-Analytica, are indeed a serious threat to democracy, despite being dismissed by some as science fiction.
In light of this, we investigated whether a supervised dimensionality-reduction approach that takes into account a host of meaningful can potentially improve the predictive performance of personality models. Such an approach could pave the way to a new family of personality models and could advance the study of personality. Alternatively, it may very well be the case that the FFM indeed “carves nature at its joints” and provides the most accurate ontology of the psychological proclivities of humans. In such a case, the FFM may remain the best predictive model of personality, and our approach will not provide improvements in predictions.
In order to examine this question, we conducted three studies. In Study 1, we built a supervised learning model using big data of personality questionnaire items and diverse, important life outcomes. We reduced the dimensionality of 100 questionnaire items into a set of five dimensions, with the objective of simultaneously minimizing prediction errors across ten meaningful life outcomes. We hypothesized that the resulting five-dimensional representation will outperform the FFM representation–when fitting a new model and attempting to predict the ten important outcomes on a held-out dataset. Next, in Studies 2 and 3, we explored the performance of the resulting model on new outcome variables.
Participants
The analyses relied on the myPersonality dataset that was collected between 2007 and 2012 via the myPersonality Facebook application. The myPersonality database is no longer shared by its creators for additional use. We received approval to download that data from the administrators of myPersonality on January 7th, 2018, and downloaded the data shortly thereafter. After the myPersonality database was taken down in 2018, we sent an email to the administrators (on June 8th, 2018), and received confirmation that we can use the data we have already downloaded. The application enabled its users to take various validated psychological and psychometric tests, such as different versions of the International Personality Item Pool (IPIP) questionnaire. Many participants also provided informed consent for researchers to access their Facebook usage details (e.g., liked pages). Participation was voluntary and likely motivated by people’s desire for self-knowledge 24 . The Participants in the myPersonality database are relatively representative of the overall population 25 . All participants provided informed consent for the data they provided to be used in subsequent psychological studies. We used data from 397,851 participants (210,279 females, 142,497 males, and 44,805 did not identify) who answered all of the questions on the 100-item IPIP representation of Goldberg’s 26 markers for the FFM which are freely available for all types of use. Participants’ mean age was 25.7 years ( SD = 8.84). The study was approved by the Institutional Review Board of Ben-Gurion University, and was conducted in accordance with relevant guidelines and regulations.
Dependent variables
We sought to use supervised learning in order to find a low-dimensional representation of personality that can be used to predict psychological consequences across a diverse set of domains. We thus focused on ten meaningful outcome variables that were available in the myPersonality database, that cover many dimensions of human life which psychologists care about:
(1) Intelligence Quotient (IQ), measured with a brief 20 items version of the Raven’s Standard Progressive Matrices test 27 .
(2) Well-being, measured with the Satisfaction with Life scale 28 .
Personal values, measured using two scores representing the two axes from the Schwartz's Values Survey:
(3) Self-transcendence vs. Self-enhancement values and
(4) Openness to Change vs. Conservation values 29 .
(5) Empathy, measured with the Empathy Quotient Scale 30 .
(6) Depression, measured with The Center for Epidemiologic Study Depression (CES-D) scale 31 .
(7) Risky behavior, measured with a single-item question concerning illegal drug use.
(8) Self-reports of legal, yet unhealthy behavior (measured as averaging two single-item questions concerning alcohol consumption and smoking).
(9) Single item self-report of political ideology.
(10) The number of friends of participants’ had on the social network Facebook.
Independent variables
Our independent variables were the participants’ answers to the 100 questions included in the IPIP-100 questionnaire 32 . In this questionnaire, the participants are asked to rate their agreement with various statements related to different behaviors in their life and their general characteristics and competencies, on a scale from 1 (strongly disagree) to 5 (strongly agree). The original use of this questionnaire is to reliably gauge participants' scores on each of the FFM dimensions. It includes five subscales, each containing 20 items; the factor score for each FFM dimension can be calculated as a simple average of these 20 questions (after reverse coding some items). In the current research we treat each item from this list of 100 questions as a separate independent variable, and seek to reduce the dimensionality of this vector using supervised learning.
Model construction
The problem we set out to solve is to find a good predictive model that is: (a) based on the 100 questions of the existing IPIP-100 questionnaire, and (b) uses five variables only, so we can fairly compare it with the FFM. Reduced Rank Regression (RRR) is a tool that allows just that: it can be used to compress the original 100 IPIP items, to a set of five new variables. These new variables are constructed so that they are good predictors, on average, of a large set of outcomes. Unlike Principal Component Analysis (PCA) or Factor Analysis, RRR reduces data dimensionality by optimizing predictive accuracy.
We randomly divided our data into an independent train and test sets. Each subject in the train and test set had 100 scores of the IPIP questionnaire ( \({X}_{1},{X}_{2},...{,X}_{100}\) ), as well as their score in each of the ten dependent variables ( \({Y}_{1},{Y}_{2},...{,Y}_{10}\) ).
X ( n × 100) and Y ( n × 100) have been centered and scaled. We fitted a linear predictor, with coefficient vector:
And in matrix notation:
Our linear predictors were fully characterized by the matrix C. We wanted these predictors to satisfy the following criteria: (a) minimize the squared prediction loss (b) consist of 5 predictors, i.e., rank ( C ) = r = 5. Criterion (a) ensures the goodness of fit of the model, and criterion (b) ensures a fair comparison with the FFM. The RRR problem amounts to finding a set of predictors, \(\hat{C}\) , so that:
where || \(\cdot \) || denotes the Frobenius matrix norm. The matrix \(C\) can be expressed as a product of two rank-constrained matrices:
where \(B\) is of has p rows and r columns, denoted, p × r , and \({A}\) is of dimension q × r . The model (2) may thus be rewritten as:
The n × r matrix \(X\hat{B}\) , which we noted \(\tilde{X}\) , may be interpreted as our new low-dimension personality representation. Crucially for our purposes, the same set of r predictors is used for all dependent variables. By choosing dependent variables from different domains, we dare argue that this set of predictors can serve as a set of “generalizable predictors”, which we call henceforth the Predictive Five (PF). For the details of the estimation of \(\hat{B}\) see the attached code. For a good description of the RRR algorithm see 33 .
Model assessment
To assess the predictive performance of the PF, and compare it to the predictive properties of the classical FFM, we used a fourfold cross validation scheme. The validation worked as follows: we learned \(\hat{B}\) from a train set (397,851 participants) using RRR; we then divided the independent test set (800 participants) into 4 subsets; we learned \(\hat{A}\) from a three-quarters part of the test set (600 participants), and computed the R 2 on the holdout test set (200 participants); we iterated this process over the 4-test subsets. The rationale of this scheme is that: (a) predictive performance is assessed using R 2 on a completely novel dataset ; (b) when learning the predictive model, we wanted to treat the personality attributes as known. We thus learned \(\hat{B}\) and \(\hat{A}\) from different sets. The size of the holdout set was selected so that R 2 estimates will have low variance. The details of the process can be found in the accompanying code ( https://github.com/GalBenY/Predictive-Five ).
To examine the performance of the RRR algorithm against another candidate reference model we also performed Principal Component Regression (PCR), where we reduced the IPIP questionnaire to its 5 leading principal components, which were then used to predict the outcome variables. We used the resulting model as a point of comparison in follow-up assessment of predictive accuracy. Like the RRR case, we learned the principal components from the train-set (397,851 participants). Next we divided the independent test set (800 participants) into 4 subsets and used a fourfold cross validation: ¾ to learn 5 coefficients, and ¼ to compute.
In order to calculate the significance of the difference in the predictive accuracy of the models we took the following approach: predictions are essentially paired, since they originate from the same participant. For each participant, we thus computed the (holdout) difference between the (absolute) error of the PF and FFM models: \(|{{\widehat{y}}_{i}}^{PF}|-|{{\widehat{y}}_{i}}^{FFM}|\) . Given a sample of such differences, comparing the models collapses to a univariate t-test allowing us to reject the null hypothesis that the mean of the differences is 0.
PF loadings
Each of the resulting PF dimensions were a weighted linear combination of IPIP-100 item responses. Despite the fact that the resulting model was based on a questionnaire meant to reliably gauge the FFM, the resulting outcome did not fully recapitulate the FFM structure. The detailed loadings for each of the resulting five dimensions appears in the supplementary materials (Fig. 1 , Supplementary Materials), can be examined in an online application we have created ( https://predictivefive.shinyapps.io/PredictiveFive ), and can be easily gleaned by examining the correlation of PF scores to the FFM scores (Fig. 2 ). None of the PF dimensions strongly correlated with demographic variables (Table 1 , Supplementary Materials). In Fig. 1 , we display the correlations between the ten outcome variables, five principle components of these outcome variables (capturing 86% of the total variance), and the five PF dimensions. For example, it can be observed the PF 3 is inversely related to performance on the intelligence test and to empathy.
Correlations between the 10 outcome variables, 5 principle components of outcome variables, and the 5 PF dimensions.
Correlations between the PF and FFM scale scores.
Predictive performance
The out-of-sample R 2 of the three models is reported in Table 1 . From this figure, we learn that the PF-based regression model is a better predictor of the outcome variables. This holds true on average (over behavioral outcomes), but also for nine of the ten outcomes individually. On 5 of the 10 comparisons, the PF-based model significantly outperformed the FFM, and in a single case the FFM-based model significantly outperformed the PF. The average improvement across all 10 measures was 40.8%.
Reproducibility analysis
If it were the case that our model discovery process produces very different loadings when run on different samples of participants, then the ontological status of the PF representation should be called into question.
In order to assess the reproducibility of the PF we split the training dataset from Study 1 into two datasets; sample A with 198,850 participants and sample B with 198,851 participants. We then learned the rotation matrix, B, on each data part, and applied it. Equipped with two independent copies of the PF, \({X}_{l }{\widehat{B}}_{l}, l=\{A,B\}\) replicability is measured by the correlation between data-parts, over participants. Table 2 reports this correlation, averaged over the 5 PFs (column “Correlation between replications”). As can be seen, the correlation between the replications is satisfactory-to-high and ranges from 0.7 to 0.98. This suggests that PF representation replicates well across samples.
Reliability analysis
If the same individuals, tested on different occasions, receive markedly different scores on the PF dimensions, then the ontological status of the PF representation should be called into question. To this end, we exploit the fact that 96,682 users answered the IPIP questionnaire twice. The test–retest correlation between these two answers is reported in Table 2 (column “Test–retest correlation”). It varied from 0.69 for the Dimension 3, to 0.79 for both Dimensions 1 and 5, suggesting that the variance captured by these dimensions is indeed (relatively) stable.
Divergence from the FFM
The superior predictive performance of the PF representation provides evidence that it differs from the FFM. Additionally, as can be gleaned from Fig. 2 (and from the detailed factor loadings’ Supplemental Material), Dimensions 3 and 4 reflect a relatively even combination of several FFM dimensions.
However, these observations do not provide us with an estimate of the degree of agreement between the two multidimensional spaces. Prevalent statistical methods of assessment of discriminant validity 34 are also not suitable to answer our question regarding the convergence\divergence between the PF and FFM spaces. These various methods only provide researchers with estimates of the agreement between unidimensional constructs .
Nonetheless, the underlying logic behind these methods (i.e., a formalization of a multitrait-multimethod matrix 35 ) is still applicable to our case. We calculated an estimate of agreement between the FFM and the PF spaces using cosine similarity , which gauges the angle between two points in a multidimensional space (the smaller the angle, the closer are the points). Our rationale is that if the FFM scores differ from the PF, they should span different spaces. The cosine similarity within measures (in our case, first and second measurements, denoted T1 and T2) should thus be larger than the similarity between measures (FFM to PF).
We used the data from the 96,682 participants for which we had test–retest data. Instead of computing standard test–retest correlations, we calculated a multidimensional test-rest score as the cosine similarity of participants’ scores on the first and second measurement, for both the FFM and PF. These estimates are expected to be highly similar and provide an upper bound on the similarity measure, partially analogous to the diameter of the multitrait-multimethod matrix. In a second stage, for each T1 and T2 vector, we measured the extent to which participants’ FFM scores are similar to their PF score, thereby calculating a magnitude that is analogous to measures of divergent validity . Because cosine similarity is sensitive to the sign and order of dimensions, we extracted the maximal possible similarity between the two spaces, providing the most conservative estimate of divergent validity.
As can be seen in Fig. 3 , the T1-T2 similarity of the FFM is nearly maximal ( M = 0.994, SD = 0.011); the T1-T2 similarity of PF is also very high ( M = 0.969, SD = 0.100). The similarity between the FFM and the PF on both T1 and T2 is much lower ( M = 0.730, SD = 0.111). The minimal difference between the convergence measures and divergence measures is on the magnitude of Hedge's g of 2.217, clearly representing a substantial divergence between the FFM and PF spaces. In other words, while the PF representation bears some resemblance, it is clearly a different representation.
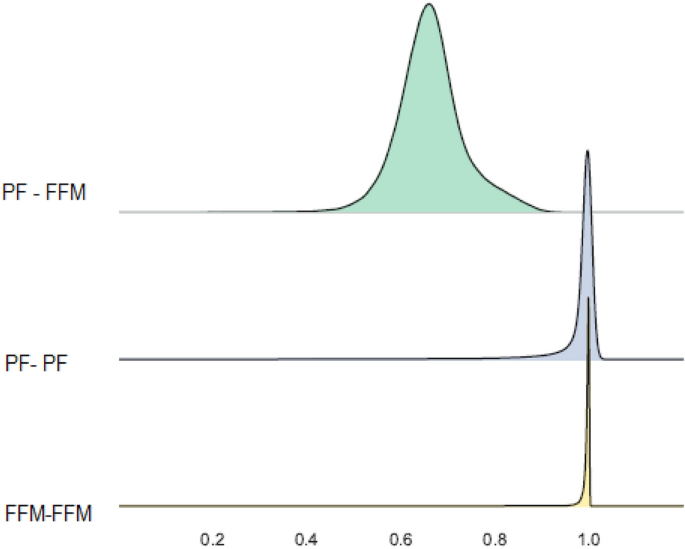
Distribution, over participants, of the multidimensional similarity between the FFM and PF representations.
The results of Study 1 provide evidence that a supervised dimensionality reduction method can yield a low-dimensional representation that is simultaneously predictive of a set of psychological outcome variables. We demonstrate that by using a standard personality questionnaire and supervised learning methods, it is possible to improve the overall prediction of a set of 10 important psychological outcomes, even when restricting ourselves to 5 dimensions of personality. RRR allowed us to compress the 100 questions of the personality questionnaire to a new quintet of attributes that optimize prediction across a large set of psychological outcomes. The resulting set of five dimensions differs from the FFM, and has better predictive power on the held-out sample than the classical FFM and an additional comparison benchmark of five dimensions generated using Principal Component Analysis.
A theory of personality should strive to predict humans’ thoughts, feelings, and behaviors across different life contexts. Indeed, the representation we discovered in Study 1 was superior to the FFM in terms of its ability to predict a diverse set of psychological outcomes on a set of novel observations. The fact that the same low-dimensional representation was applicable across a set of important outcomes of human psychology suggests that it is a relatively generalizable model, in the sense that it simultaneously applies to several important domains. However, despite the diversity of the outcome measures examined in Study 1, it remains possible that the PF representation is only effective for the prediction of the set of outcome measures on which it was trained. Such a finding would not negate the usefulness of this model, given the wide variety of outcomes captured by the PF. However, it is interesting to see whether the resulting representation can improve prediction on additional sets of outcomes. In light of this, in Study 2 we sought to examine the performance of the PF on a set of novel outcome measures that were present in the myPersonality database, but that were held-out from the model generation process. Specifically, in this study we sought to see whether the PF representation outperforms the FFM in its ability to predict participants’ experiences during their childhood .
Unlike the outcome measures used in Study 1, this dependent variable does not pertain to participants’ lives in the present, rather, it is a measure of their past experiences. As such, “retrodiction” of remote history may be especially challenging. Nonetheless, it is widely held that individuals’ psychological properties are shaped, at least to some extent, by the degree to which they were raised in a loving household 36 , 37 . Indeed, there is evidence to the fact that many specific psychological attributes are shaped by experiences with primary caregivers (e.g., shared environmental effects on topics such as food preference 38 , substance abuse 39 , and agression 40 ). In light of this, we reasoned that it is reasonable to expect that one's personality profile should contain information that is predictive of individuals' retrospective reports of their upbringing.
We used data from 3869 participants who answered all of the questions on the 100-item IPIP representation of 26 markers for the Big Five factor structure, and answered the short form “My Memories of Upbringing” (EMBU) questionnaire 41 .
The short form of the EMBU includes a total of six subscales: three subscales that contain questions to measure the extent to which the participants' father was a warm , rejecting , and overprotecting parent, and three subscales that measure the extent to which the participants' mother was warm , rejecting , and overprotecting .
As can be seen in Table 1 , for all six variables, prediction accuracy was relatively low; however, importantly, in all six cases the PF-based model outperformed the FFM-based model, and was significantly better for four out of the six outcome variables. The average improvement across the six outcome measures was 49.2%.
The results of Study 2 further support the idea that the PF representation that was built using the 10 meaningful outcome measures present in the myPersonality database is at least somewhat generalizable. However, Study 2 again relied on myPersonality participants, upon which the PF was built. In light of this, in Study 3 we sought to further test the generality of the PF by examining whether it outperforms the FFM-based model on a set of new participants. Furthermore, we wanted to see whether our model can outperform the FFM-based model on a set of new outcome measures selected by an independent group of professional psychologists, blind to our model-generation procedure.
We collected new data using Amazon’s Mechanical Turk ( www.MTurk.com ). M-Turk is an online marketplace that enables data collection from a diverse workforce who are paid upon successful completion of each task. Our target sample size was 500 participants, which is double the size of what is considered a standard, adequate sample size in individual differences research 42 . In practice, 582 participants participated in the study, 35 of them were omitted for failing attention checks, leaving 547 participants in the final dataset (243 females and 304 men). This number exceed a sample size of 470 participants which provides 95% confidence that a small effect (⍴ = 0.1) will be estimated with narrow (w = 0.1) Corridor of Stability 42 .
In order to make sure that the PF generalize across different domains of psychological interest, it was important to generate the list of outcome variables in a way that is not biased by our knowledge of the original ten outcome variables on which the PF was designed (i.e., intelligence, well-being, and so on). Therefore, on January 3rd, 2019, we gathered a list of 12 new outcome measures by posting a call on the Facebook group PsychMAP ( https://www.facebook.com/groups/psychmap ) asking researchers: “to name psychological outcome measures that you find interesting, important, and that can be measured on M-Turk using a single questionnaire item on a Likert scale.” Once we arrived at the target number of questions we closed the discussion and stopped collecting additional variables. The 12 items were suggested by eight different psychologists, six of which had a PhD in psychology and five were principal investigators. By using this variable elicitation method, we had no control over the outcome measures, and could be certain that we have gathered a randomly-chosen sample of outcomes that are of interest to psychologists.
This arbitrariness of the outcome generation process (selecting the first 12 outcomes nominated by psychologists, without any consideration of consensus views regarding variable importance)—and the likely low psychometric reliability of single-item measures–can be seen as a limitation of this study. However, our reasoning was that such a situation best approximates the "messiness" of the unexpected, noisy, real-world scenarios wherein prediction may be of interest–and as such, provides a good test of predictive performance of the FFM and PF.
In the M-turk study, participants rated their agreement with 12 statements (1- Strongly Disagree to 7- Strongly Agree). The elicited items were:
(1) “I care deeply about being a good person at heart”.
(2) “I value following my heart/intuition over carefully reasoning about problems in my life”.
(3) “Other people's pain is very real to me”.
(4) “It is important to me to have power over other people”.
(5) “I have always been an honest person”.
(6) “When someone reveals that s/he is lonely I want to keep my distance from him/her”.
(7) “Before an important decision, I ask myself what my parents would think”.
(8) “I have math anxiety”.
(9) “I am typically very anxious”.
(10) “I enjoy playing with fire”.
(11) “I am a hardcore sports fan”.
(12) “Politically speaking, I consider myself to be very conservative”.
The independent variables were participants’ answers to the 100 questions of the IPIP questionnaire.
Similarly to Study 1, we use a fourfold cross validation scheme in order to assess the predictive performance of the PF on new data set and outcome variables. Next, we compared it to the predictive performance of the FFM. The validation worked as follows: we had \(\hat{B}\) from Study 1, we learned \(\hat{A}\) from a part of the new sample (400 ~ participants) and computed the R 2 on the holdout test set (130 ~ participants). In the spirit of the fourfold cross-validation, we iterated this process over the 4-test sets and calculated the average test R 2 for each model.
Similarly to Studies 1–2, the results showed that the predictive performance of the PF was again better than that of the Big Five, although the improvements were more modest (average 30% improvement across the 12 measures). In 5 out of 12 cases, the PF-based model was significantly better than the FFM-based model, and the opposite was true in 2 cases.
The out-of-sample R 2 of the two models (PF\Big Five) in Study 3 show a consistent trend with the results presented earlier in Study 1 and Study 2, that is, a somewhat higher percentage of explained variance in the models with the PF as predictors. This improvement observed in Study 3 was more modest than that observed earlier, but is nonetheless non-trivial—given that the set of outcome variables was different from the one the PF representation was trained on, and given that the PF representation was trained on items from questionnaires designed to measure the FFM. As such, the results of Studies 1–3 clearly demonstrate the generalizability of the PF.
A potential criticism of these findings is that the success of the PF model was more prominent on variables that were more similar to the 10 dependent measures upon which the PF was trained. However, it is important to keep in mind that the 12 outcome measures in this study were selected at random by an external group of psychologists. As such, this primarily means that the 10 psychological outcomes used to train the PF indeed provide good coverage of psychological processes that are of interest to psychologists, and thereby, overall, generalize well to novel prediction challenges.
General discussion
In this contribution, we set out to examine the viability of a novel approach to modeling human personality. Unlike the prevailing Five-Factor Model (FFM) of personality, which was developed by relying on unsupervised dimensionality reduction techniques (i.e., Factor Analysis), we utilized supervised machine learning techniques for dimensionality reduction, using numerous psychologically meaningful outcomes as data labels (e.g., intelligence, well-being, sociability). Whereas the FFM is optimized towards discovering an ontology that explains most of the variance on self-report measures of psychological traits, our new approach devised a low-dimensional representation of human trait statements that is optimized towards prediction of life outcomes. Indeed, the results showed that our model, which we term the Predictive Five (PF), provides predictive performance that is better than the one achieved by the FFM in independent validation datasets (Study 1–2), and on a new set of outcome variables, selected independently of the first study (Study 3). The main contribution of the current work is explicating and demonstrating a methodological approach of generating a personality representation. However, the results of this work is a specific representation that is of interest and of potential use in and of itself. We now turn to discuss both our general approach and the resulting representation.
Interpreting the PF
The dimensional structure that emerged when using our supervised-dimensionality reduction approach differed from the FFM. Two dimensions (Dimension 1 and 2) largely reproduced the original FFM factors of Extraversion and Neuroticism. Interestingly, these two dimensions are the ones that were highlighted in early psychological research as the “Big Two” factors of personality (Wiggins, 1966). Dimension 5 was also highly related to an existing FFM dimension, namely, Openness to Experience .
The third and fourth dimensions in the model did not correspond to a single FFM trait, but were composed of a mixture of various items. An inspection of the loadings suggests that Dimension 4 is related to some sort of a combative attitude, perhaps captured best by the construct of Dominance 43 , 44 , 45 . The items that loaded highly on this dimension related to hostility (“Do not sympathize with others”; “Insult people”), a right-wing political orientation (“Do not vote for liberal political candidates”), and an approach-oriented 46 stance (“Get chores done right away”; “Find it easy to get down to work”).
Like PF Dimension 4, Dimension 3 also seemed to capture approach-oriented characteristics (with high loadings for the items “Get chores done right away” and “Find it easy to get down to work”), however, this dimension differed from Dimension 4 in that it represented a harmony-seeking phenotype 47 . The items highly loaded on this dimension were those associated with low levels of narcissism (“keep in the background”, “do not believe I am better than others”) but with a stable self-worth (“am pleased with myself”). Additional items that were highly loaded on this dimension were those that reflect cooperativity (“concerned with others” and “sympathize with others”).
These two dimensions may seem like dialectical opposites. Indeed, the item “sympathize with others” strongly loaded on both factors, but with a different sign. However, the additional items that strongly loaded on these two dimensions appear to have provided a context that altered the meaning of this item. This is evident in the fact that Dimensions 3 and 4 are not correlated with each other. A possible speculative interpretation is that the two phenotypes captured by Dimensions 3 and 4 can be thought of as two strategies that may have been adaptive throughout human evolution. The first, captured by Dimension 4 seems to represent aggressive traits that may have been especially useful in the context of inter -group competition and conflict; the second, captured by Dimension 3, seems to represent traits that may be associated with intra -group cooperation and peace.
In general, the interpretability of the PF representation is lower than that of the FFM, with some surprising items loaded together on the same dimension. For example, the two agreeableness items that “do not believe I am better than others” and “respect others” that are strongly correlated with each other were highly loaded onto Dimension 1 (that is related to introversion), but with opposite signs. To a certain extent, this is a limitation of the predictive approach in psychology. However, such confusing associations may lead us towards identifying novel insights. For example, it is possible that some individuals adopt an irreverent stance towards both self and others, and such a stance could be predictive of various psychological outcomes, and correlated with introversion.
Towards a more predictive science of personality
As noted, the reasons that people seek models of personality are twofold: first, we want models that allow us to understand, discuss and study the differences between people; second, we need these models in order to be able to predict and affect people’s choices, feelings and behaviors 48 . Current approaches to personality modeling succeeded on the former, providing highly comprehensible dimensions of individual differences (e.g., we can easily understand and communicate the contents of the dimension of “Neuroticism” by using this sparse semantic label). However, the ability of the FFM to accurately predict outcomes in people’s lives is at least somewhat limited 19 , 20 , 20 , 49 .
The significance of the current work is that it describes a new approach to modeling human personality, that makes the prediction of behavior an explicit and fundamental goal. Our research shows that supervised dimensionality reduction methods can generate relatively generalizable, low-dimensional models of personality with somewhat improved predictive accuracy. Such an approach could complement the unsupervised dimensionality reduction models that have prevailed for decades in personality research. Moreover, this research can complement attempts to improve the predictive validity of psychology by using non-parsimonious (i.e., facets and item-level) questionnaire-based predictive models 50 .
Aside from providing a general approach for the generation of personality models, the current research also provides a potentially useful instrument for psychologists across different domains of psychological investigation. Our findings suggest that psychologists who are interested in predicting meaningful consequences (e.g., workplace or romantic compatibility) or in optimizing interventions on the basis of individuals’ characteristics (e.g., finding out which individuals will best respond to a given therapeutic technique)—may benefit from incorporating the PF dimensions in their predictive models. To facilitate such future research, we provide the R code that calculates the five dimensions based on answers on the freely available IPIP-100 questionnaire ( https://github.com/GalBenY/Predictive-Five ). The use of an existing, open-access, widely-used questionnaire means that researchers can now easily apply the PF coding scheme alongside with the FFM coding scheme to their data, and compare the utility of the two models in their own specific research domains.
One avenue of potential use of the PF representation is in clinical research. The PF showed improved prediction of depression and well-being; moreover, the PF substantially outperformed the FFM in the prediction of two known resilience factors (intelligence and empathy). Specifically, PF Dimension 3 (which, as noted above, seems to represent some harmony-seeking phenotype) significantly contributed to the prediction of all of four outcomes. As such, future work could further investigate the incremental validity of this dimension (and the PF representation more generally) as a global resilience indicator.
Across a set of 28 comparisons, the predictions derived from the PF-based model were significantly better in 15 cases, and significantly worse in 3 cases. The average improvement in R 2 across the 28 outcomes was 37.7%. However, it is important to note that the PF representation described herein is just a first proof of concept of this general approach, and it is likely that future attempts that are untethered to the constraints undertaken in the current study can provide models of greater predictive accuracy. Specifically, in the current research we relied on the IPIP-100, a questionnaire designed by researchers specifically in order to reliability measure the factors of the FFM, and limited ourselves to a five-dimension solution, to allow comparison with the FFMs. The PF representation outperformed the FFM representation despite these constraints. These results provide a very conservative test for the utility of our approach.
Future directions
Future attempts to generate generalizable predictive models will likely produce even stronger predictive performance if they relax the constraint of finding exactly five dimensions and perform dimensionality-reduction based on the raw data used to generate the FFM itself—namely, the long list of trait adjectives that exist in human language, and that were reduced into the five dimensions of the FFM.
For the sake of simplicity comparability to the FFM, the current work employed a linear method for supervised dimensionality reduction. Recent work in machine learning has demonstrated the power of Deep Neural Networks as tools for dimensionality reduction (e.g., language embedding models). In light of this, it is likely that future work that utilizes non-linear methods for supervised dimensionality reduction could generate ever more predictive representations (i.e., “personality embeddings”).
A limitation of the current work is that the PF was trained on a relatively limited set of 10 important life outcomes (e.g., IQ, well-being, etc.). While these outcome measures seem to cover many of the important consequences humans care about (as evident by the predictive performance on Study 3), it is likely that training a PF model on a larger set of outcome variables will improve the coverage and generalizability of future (supervised) personality models. A potential downside of extending the set of outcome measures used for training, is that at some point (e.g., 20, 100 outcomes) it is possible that the “blanket will become too short”: namely, that it will be difficult to find a low-dimensional representation that arrives at satisfactory prediction performance simultaneously across all outcomes. Thus, future research aiming at generating more predictive personality models may need to find a “sweet spot” that allows the model to fit to a sufficiently comprehensive array of target outcomes.
What may be the most important consequence of the current approach is that whereas previous attempts of modeling human personality necessarily limited by their reliance on the subjective products of the human mind (i.e., were predicated on human-made psychological theories, or subjective ratings of trait words), our approach holds the unique potential of generating personality representations that are based on objective inputs.
A final question concerning predictive models of personality is whether we even want to generate such models, given the potential of their misuse. While the current results still show the majority of variance in psychological outcomes remain unexplained–in the era of social networks and commercial genetic testing, the predictive approach to personality modeling could theoretically lead to models that render human behavior highly predictable. Such models give rise to both ethical concerns (e.g., unethical use by governments and private companies, as in the Cambridge-Analytica scandal) and moral qualms (e.g., if behavior becomes highly predictable, what will it mean for notions of free will and personal responsibility?). While these are all valid concerns, we believe that like all other scientific advancements, personality models are tools that can provide a meaningful contribution to human life (e.g., predicting suicide in order to avoid it; predicting which occupation will make a person happiest). As such, the important, inescapable quest towards generating even more effective models that will allow us to predict and intervene in human behavior is only just the beginning.
Data availability
The data for Study 1, 3 and 4 rely on the myPersonality database ( www.mypersonality.org ) which is an unprecedented big-data repository for psychological research, used in more than a hundred publications. We achieved permission from the owners of the data to use it for the current research—but we do not have their permission to share it for wider use. The data for Study 2 is available upon request. We also share the complete code and the full model with factor loadings ( https://github.com/GalBenY/Predictive-Five ).
Newcomb, T. & Heider, F. The psychology of interpersonal relations. Am. Sociol. Rev. 23 , 742 (1958).
Article Google Scholar
Dweck, C. S. Self-Theories: Their Role in Motivation, Personality, and Development (Psychology press, London, 2013).
Book Google Scholar
Swann, W. B. Jr. Quest for accuracy in person perception: A matter of pragmatics. Psychol. Rev. 91 , 457–477 (1984).
Ægisdóttir, S. et al. The meta-analysis of clinical judgment project: Fifty-six years of accumulated research on clinical versus statistical prediction. Couns. Psychol. 34 , 341–382 (2006).
Allport, G. W. & Odbert, H. S. Trait-names: A psycho-lexical study. Psychol. Monogr. 47 , i (1936).
Costa, P. T. Jr. & McCrae, R. R. Personality: Another ‘hidden factor’ is stress research. Psychol. Inq. 1 , 22–24 (1990).
Allik, I. & Allik, I. U. The Five-Factor Model of Personality Across Cultures (Springer, Berlin, 2002).
Google Scholar
Benet-Martínez, V. & John, O. P. Los Cinco Grandes across cultures and ethnic groups: Multitrait-multimethod analyses of the Big Five in Spanish and English. J. Pers. Soc. Psychol. 75 , 729–750 (1998).
Laajaj, R. et al. Challenges to capture the big five personality traits in non-WEIRD populations. Sci. Adv. 5 , eaaw5226 (2019).
Article ADS Google Scholar
John, O. P. The ‘Big Five’ factor taxonomy: Dimensions of personality in the natural language and in questionnaires. Handbook of personality: Theory and research (1990).
McCrae, R. R. & Costa, P. T. Clinical assessment can benefit from recent advances in personality psychology. Am. Psychol. 41 , 1001–1003 (1986).
John, O. P. & Srivastava, S. The Big Five trait taxonomy: History, measurement, and theoretical perspectives. Handbook of personality: Theory and (1999).
Gilead, M., Trope, Y. & Liberman, N. Above and beyond the concrete: The diverse representational substrates of the predictive brain. Behav. Brain Sci. 43 , e121 (2019).
Chen, L. Curse of dimensionality. Encycl. Database Syst. https://doi.org/10.1007/978-1-4614-8265-9_133 (2018).
Kosinski, M., Stillwell, D. & Graepel, T. Private traits and attributes are predictable from digital records of human behavior. Proc. Natl. Acad. Sci. USA 110 , 5802–5805 (2013).
Article ADS CAS Google Scholar
Confessore, N. Cambridge Analytica and Facebook: The Scandal and the Fallout So Far. The New York Times (2018).
Harari, Y. N. Homo Deus: A Brief History of Tomorrow (Random House, London, 2016).
Hough, L. M. The ‘Big Five’ personality variables-construct confusion: Description versus prediction. Hum. Perform. 5 , 139–155 (1992).
Sibley, C. G., Osborne, D. & Duckitt, J. Personality and political orientation: Meta-analysis and test of a threat-constraint model. J. Res. Pers. 46 , 664–677 (2012).
Morgeson, F. P. et al. Are we getting fooled again? Coming to terms with limitations in the use of personality tests for personnel selection. Pers. Psychol. 60 , 1029–1049 (2007).
Gibney, E. The scant science behind Cambridge Analytica’s controversial marketing techniques. Nature https://doi.org/10.1038/d41586-018-03880-4 (2018).
Article PubMed Google Scholar
Matz, S. C., Kosinski, M., Nave, G. & Stillwell, D. J. Psychological targeting as an effective approach to digital mass persuasion. Proc. Natl. Acad. Sci. USA 114 , 12714–12719 (2017).
Article CAS Google Scholar
Roberts, B. W., Kuncel, N. R., Shiner, R., Caspi, A. & Goldberg, L. R. The power of personality: The comparative validity of personality traits, socioeconomic status, and cognitive ability for predicting important life outcomes. Perspect. Psychol. Sci. 2 , 313–345 (2007).
Kosinski, M., Matz, S. C., Gosling, S. D., Popov, V. & Stillwell, D. Facebook as a research tool for the social sciences: Opportunities, challenges, ethical considerations, and practical guidelines. Am. Psychol. 70 , 543–556 (2015).
Stillwell & Kosinski. myPersonality project: Example of successful utilization of online social networks for large-scale social research. Am. Psychol.
Goldberg, L. R. The development of markers for the Big-Five factor structure. Psychol. Assess. 4 , 26–42 (1992).
Stillwell, D. J. & Kosinski, M. myPersonality Project website. (2015).
Diener, E. D., Emmons, R. A., Larsen, R. J. & Griffin, S. The satisfaction with life scale. J. Pers. Assess. 49 , 71–75 (1985).
Schwartz, S. H. Universals in the content and structure of values: Theoretical advances and empirical tests in 20 countries. In Advances in Experimental Social Psychology Vol. 25 (ed. Zanna, M. P.) 1–65 (Academic Press, New York, 1992).
Baron-Cohen, S. & Wheelwright, S. The empathy quotient: an investigation of adults with Asperger syndrome or high functioning autism, and normal sex differences. J. Autism Dev. Disord. 34 , 163–175 (2004).
Radloff, L. S. The CES-D Scale: A self-report depression scale for research in the general population. Appl. Psychol. Meas. 1 , 385–401 (1977).
Cucina, J. M., Goldenberg, R. & Vasilopoulos, N. L. Confirmatory factor analysis of the NEO-PI-R equivalent IPIP inventory. PsycEXTRA Dataset https://doi.org/10.1037/e518612013-349 (2005).
Chen, L. Sparse Reduced-Rank Regression for Simultaneous Dimension Reduction and Variable Selection in Multivariate Regression.
Henseler, J., Ringle, C. M. & Sarstedt, M. A new criterion for assessing discriminant validity in variance-based structural equation modeling. J. Acad. Mark. Sci. 43 , 115–135 (2015).
Campbell, D. T. & Fiske, D. W. Convergent and discriminant validation by the multitrait-multimethod matrix. Psychol. Bull. 56 , 81–105 (1959).
Winnicott, D. W. The Child, the Family, and the Outside World: By DW Winnicott (Penguin Books, London, 1969).
Bowlby, J. A Secure Base: Clinical Applications of Attachment Theory (Taylor & Francis, New York, 2005).
Smith, A. D. et al. Food fussiness and food neophobia share a common etiology in early childhood. J. Child Psychol. Psychiatry 58 , 189–196 (2017).
Verhulst, B., Neale, M. C. & Kendler, K. S. The heritability of alcohol use disorders: A meta-analysis of twin and adoption studies. Psychol. Med. 45 , 1061–1072 (2015).
Chen, J., Yu, J., Zhang, J., Li, X. & McGue, M. Investigating genetic and environmental contributions to adolescent externalizing behavior in a collectivistic culture: A multi-informant twin study. Psychol. Med. 45 , 1989–1997 (2015).
Arrindell, W. A. et al. The development of a short form of the EMBU: Its appraisal with students in Greece, Guatemala, Hungary and Italy. Pers. Individ. Dif. 27 , 613–628 (1999).
Schönbrodt, F. D. & Perugini, M. At what sample size do correlations stabilize?. J. Res. Pers. 47 , 609–612 (2013).
Murray, H. A. Explorations in personality: a clinical and experimental study of fifty men of college age. 761 , (1938).
Jackson, D. N. Personality research form (Research psychologists Press, 1965).
Pratto, F., Sidanius, J., Stallworth, L. M. & Malle, B. F. Social dominance orientation: A personality variable predicting social and political attitudes. J. Pers. Soc. Psychol. 67 , 741–763 (1994).
Higgins, E. T., Kruglanski, A. W. & Pierro, A. Regulatory mode: Locomotion and assessment as distinct orientations. In Advances in Experimental Social Psychology Vol. 35 (ed. Zanna, M. P.) 293–344 (New York, Elsevier Academic Press, 2003).
Leung, K., Koch, P. T. & Lu, L. A dualistic model of harmony and its implications for conflict management in Asia. Asia Pac. J. Manag. 19 , 201–220 (2002).
Saucier, G. & Srivastava, S. What makes a good structural model of personality? Evaluating the Big Five and alternatives. APA Handbook Person. Soc. Psychol. 4 , 283–305 (2015).
Salgado, J. F. The big five personality dimensions and counterproductive behaviors. Int. J. Select. Assess. 10 , 117–125 (2002).
Stewart, R. D., Mõttus, R., Seeboth, A., Soto, C. J. & Johnson, W. The finer details? The predictability of life outcomes from Big Five domains, facets, and nuances. J. Pers. 90 , 167–182 (2022).
Download references
Acknowledgements
The study was supported by ISF grant 1113/18 to M.G.
Author information
Authors and affiliations.
Department of Industrial Engineering and Management, Ben-Gurion University of the Negev, Beersheba, Israel
Pagaya Technologies, Tel Aviv, Israel
Jonathan Rosenblatt
School of Psychological Sciences and Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel
Michael Gilead
You can also search for this author in PubMed Google Scholar
Contributions
G.L., J.D.R., and M.G. conducted this work jointly.
Corresponding authors
Correspondence to Jonathan Rosenblatt or Michael Gilead .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary information., rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Lavi, G., Rosenblatt, J. & Gilead, M. A prediction-focused approach to personality modeling. Sci Rep 12 , 12650 (2022). https://doi.org/10.1038/s41598-022-16108-3
Download citation
Received : 19 December 2021
Accepted : 05 July 2022
Published : 25 July 2022
DOI : https://doi.org/10.1038/s41598-022-16108-3
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

Browse Subjects
- Intelligence service Methodology.">Methodology.
- International relations Forecasting.">Forecasting.
- Terrorism Forecasting.">Forecasting.
- World politics Forecasting.">Forecasting.
- Reference Manager
- Simple TEXT file
People also looked at
Original research article, a significant wave height prediction method based on deep learning combining the correlation between wind and wind waves.

- 1 College of Computer Science and Technology, China University of Petroleum, Qingdao, China
- 2 Department of Artificial Intelligence, Faculty of Computer Science, Polytechnical University of Madrid, Madrid, Spain
- 3 DAMO Academy, Alibaba Group, Hangzhou, China
- 4 Key Laboratory of Environmental Change and Natural Disaster of Ministry of Education, Beijing Normal University, Beijing, China
- 5 State Key Laboratory of Tropical Oceanography, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
Accurate wave height prediction is significant in ports, energy, fisheries, and other offshore operations. In this study, a regional significant wave height prediction model with a high spatial and temporal resolution is proposed based on the ConvLSTM algorithm. The model learns the intrinsic correlations of the data generated by the numerical model, making it possible to combine the correlations between wind and wind waves to improve the predictions. In addition, this study also optimizes the long-term prediction ability of the model through the proposed Mask method and Replace mechanism. The experimental results show that the introduction of the wind field can significantly improve the significant wave height prediction results. The research on the prediction effect of the entire study area and two separate stations shows that the prediction performance of the proposed model is better than the existing methods. The model makes full use of the physical correlation between wind and wind waves, and the validity is up to 24 hours. The 24-hour forecast R² reached 0.69.

1 Introduction
Wind waves are waves generated by and influenced by the local wind ( Barnett and Kenyon, 1975 ). It is characterized by often sharp wave crests, very irregular distribution on the sea surface, short crest lines, and minor periods. When the wind is strong, the phenomenon of breaking waves often occurs, and water splashes are formed. In general, wind disturbance of the sea surface causes capillary waves (ripples) so that the wind further provides the necessary roughness for delivering energy to the sea surface. Then, the waves continue to be fueled by the pressure of the wind on its surface ( Longuet-Higgins, 1963 ; Kirby, 1985 ), causing the wind waves to grow ( Phillips, 1957 ). Wind waves dominate the motion of the sea for a short period. Therefore the study of wind waves has implications for many applications such as navigation safety and coastal engineering. Waves also define air-sea fluxes and interact strongly with surface currents, upper ocean turbulence, and sea ice. Understanding and accurately predicting waves are very beneficial to humans.
In the past few decades, researchers have made great strides in studying the causes of wind waves and the correlation between wind and waves ( Barnett, 1968 ). In order to analyze the wind waves field, Sverdrup and Munk (1947) first used an empirical or semi-analytical approach. However, the method has obvious limitations ( Kamranzad et al., 2011 ). Hasselmann (1968) has also studied the evolution of the wind waves’ power spectrum in-depth and demonstrated a strong correlation between both wind and wind waves.
At this stage, the mainstream forecasting idea for oceanographers to forecast wind waves is to use numerical models. The numerical model uses oceanic elements such as wind as input and solves complex equations to produce wave forecasts. The most widely used models include the National Weather Service’s (NWS) WaveWatch III (WW3) ( Tolman et al., 2009 ), Simulating Waves Nearshore (SWAN) ( Booij et al., 1999 ) developed by the Delft University of Technology, etc ( Zheng et al., 2016 ). Traditional numerical model forecasting methods combine the advantages of physical simulation and data-driven approaches to make forecasts with high spatial and temporal resolution ( Wei et al., 2013 ). This hybrid approach of physical simulation and data-driven prediction is theoretically sound. However, it has significant limitations in practical offshore industry applications: the time lag and its accuracy cannot be guaranteed. In addition, the expensive computational and maintenance costs of the numerical model make it a prudent consideration as an operational application ( Song et al., 2022 ).
In recent years, the application of Artificial Intelligence (AI) in marine and atmospheric sciences has developed rapidly ( Van Aartrijk et al., 2002 ; Bolton and Zanna, 2019 ). AI can naturally process many data sources, such as numerical forecast results, radar, satellite, station observations, and even decision data (natural language), which is almost impossible for existing coupled sea-air numerical models. Some studies have even found that AI models outperform existing numerical models for short-term wind waves prediction ( James et al., 2018 ). Berbić et al. (2017) ; Callens et al. (2020) predicted the significant wave height within 3-hour accurately using Random Forest (RF) and Support Vector Machine (SVM), respectively. Fan et al. (2020) used the Long Short Term Memory Network(LSTM) algorithm to predict the significant wave height of several stations for 6-hour, and the results were satisfactory. Song et al. (2020) uses merged-LSTM to mine the hidden patterns in short time series to solve the long-term dependence of series variability and to make compelling predictions of sea surface height anomaly(SSHA). Meng et al. (2021) proposes a bi-directional gated recurrent unit (BiGRU) network for predicting wave heights during tropical cyclones (TCs). Artificial intelligence has the advantage of solid data drive and a high potential for model optimization, which can theoretically solve the “costly” problem of numerical forecast models while improving the “accuracy” of forecasts.
Although the application of AI in wave height prediction is becoming more and more widespread, most of them are limited to single-site forecasting. However, wind wave fields are two-dimensional fields, so predicting wave height at a point is not only a matter of time series but should also consider the spatial correlation with other surrounding points ( Jönsson et al., 2003 ; Gavrikov et al., 2016 ). In addition, most current AI applications for predicting wave height use single-factor forecasting, treating each ocean variable individually, which ignores the correlation between different ocean elements and lacks physical meaning ( Fu et al., 2019 ). Only the wave field factor is applied to forecast the wave field. The physical correlation between wind and wind waves is ignored. Zhou et al. (2021) established A two-dimensional SWH prediction model based on convolution Long and Short-term memory (ConvLSTM). However, the model only considers the wave and ignores the influence of wind. The mean absolute percentage errors of 6-hour, 12-hour, and 24-hour advance are 15%, 29%, and 61%, respectively. Moreover, the spatial and temporal resolution of the data should also be considered if the deep learning approach is to be truly applied to the problem of forecasting ocean elements. With the deep development of ocean research, human production life increasingly needs to understand the ocean elements with high spatial and temporal resolution. Most of the current deep learning wave forecasting methods are limited to low spatial and temporal resolution conditions, and such research can no longer meet the practical needs of society.
In this study, a deep learning model based on ConvLSTM was developed to combine the correlation between wind and wind waves to predict significant wave heights with high spatial and temporal resolution in the Beibu Gulf. ConvLSTM has been successfully applied to 2D precipitation prediction ( Shi et al., 2015 ). It enables the model to learn the spatial correlation of elements through a unique convolution method, which solves the problem of spatial information loss in traditional LSTM and improves the accuracy of 2D predictions. Specific modifications to the model were made in this study to enable the model to be adapted to the study sea area and to learn the correlation between wind and significant wave heights in the numerical model data. We then set up a series of experiments to evaluate the performance and accuracy of the model. The model successfully predicts the hourly significant wave height of 1/40° and has an excellent long-term prediction ability. After the model is trained, it is only necessary to provide the model with the corresponding wind speed and significant wave height data to obtain the required predicted significant wave height.
The rest of the paper is organized as follows: Section.2 describes the data and research area we used, and Section.3 describes the method used and the construction and evaluation metrics of the proposed model. Section.4 shows the predictive performance of the proposed model and corrects the problems in the prediction process. Finally, we conclude and discuss future research recommendations in Section.5.
2 Study area and data
2.1 study area.
In this work, the study area is the Beibu Gulf and its adjacent waters in the South China Sea (16°N - 23°N, 105°E - 113°E), as shown in the black box in Figure 1 . It includes the shelf waters as well as other waters around Hainan Island; the water depth gradually deepens from the shore to the central part, with an average depth of 42 meters and a maximum depth of more than 100 meters ( Gao et al., 2015 ). The study area is mainly surrounded by some cities in Guangxi, Guangdong, Hainan Province (China), and Vietnam, which are important ports and good fishing grounds ( Koongolla et al., 2020 ). The Beibu Gulf is located in tropical and subtropical areas ( Cooke et al., 2011 ). In winter, it is influenced by cold air from mainland China, with northeast winds and sea surface temperature of about 20C. In summer, the wind comes from the tropical ocean, mainly from the southwest, and the sea surface temperature is as high as 30C. It is often attacked by typhoons. Generally, about five typhoons ( Shao et al., 2018 ) pass here every year.
Figure 1 (A) South China Sea and the Beibu Gulf, and (B) Beibu Gulf and its adjacent waters.
The data used in this study are the significant wave height (SWH) and wind speed (WS) data. It is worth noting that the significant wave height data we use refers specifically to the significant wave height of wind waves. These data were provided by the South China Sea Institute of Oceanography, Chinese Academy of Sciences. These data are the products of the WAVEWATCH III and COAM models. The researchers involved have adopted the latest wind stress calculation scheme based on the third-generation wave model WW3, which improves the model’s prediction of wind and waves generated by different wind speeds and wind field variations. The model allows a better simulation of the temporal variation of the waves, and the values obtained are closer to the observed values than in the ERA5 reanalysis. The model can provide hourly forecasts with a spatial resolution of 1/40°*1/40°. A more detailed description of the data is available here ( Li et al., 2021 ). Due to the high accuracy of these data, they can be used as an approximation of the observed data in the case of insufficient actual measurement data. Since it is difficult to obtain actual measurement data with high accuracy in the study area, we used the above data as a comparison value in our study. In this study, SWH and WS data with Spatio-temporal resolution of 1h and 1/40°*1/40° for two years from 2018-2019 were selected, with 80% of the data used as the training set, 10% for validation, and 10% for testing. The maximum significant wave height in the data is 7.34m and the top wind speed is 17m/s. In Section 4, we also compare the predictions with the ERA5 reanalysis information used, which can be found here ( www.ecmwf.int/en/forecasts/datasets/reanalysis-datasets/era5 ). It is worth stating that the high-resolution data used in this study will be open-sourced to facilitate researchers in studying important wave height issues at high resolution. These data are available here: citep https://doi.org/10.5281/zenodo.6402321 .
This study focuses on the significant wave height variation over the entire study area rather than on specific stations. Therefore, for each point in the study area, we need to consider it in terms of time series and spatial relationships. This study proposes a novel prediction method based on Mask-ConvLSTM deep learning network and Replace mechanism. In this study, specific modifications are made to the ConvLSTM model, which allows the model to be adapted to our study sea area and learn the physical correlation between wind speed and significant wave height. The model was used to predict the SWH conditions after a few hours. The number of layers of the network is three, containing 6,12,2 convolutional kernels, respectively, the size of these convolutional kernels is set to 3*3, and the step size of each move is 1. Our experiments were conducted on a cluster of computers. This study used an NVIDIA TeslaV100S and Intel(R) Xeon(R) Silver 4214R CPU in terms of hardware. Regarding software, this study used Tensorflow-2.4.1 and CentOS 7.6.
3.1 Convolutional LSTM network
The LSTM algorithm, known as Long short-term memory, was first proposed by Hochreiter and Jürgen Schmidhuber in 1997 and is a particular form of RNN (Recurrent neural network), while RNN is a general term for a series of neural networks capable of processing sequential data ( Hochreiter and Schmidhuber, 1997 ). In 2005, Alex Graves and Jürgen Schmidhuber proposed a bidirectional long short-term memory neural network (BLSTM) based on LSTM, also known as vanilla LSTM ( Graves and Schmidhuber, 2005 ). It is one of the most widely used LSTM models at present. The ability of LSTM to remove or add information to nodes to change the information flow state relies on the careful regulation of the gate structure ( Meng et al., 2022 ). Gates are nodes that can be selected to pass information, and they consist of Sigmoid complexes and point-by-point multiplication operations. The ingenuity of the LSTM lies in the addition of input gates, forgetting gates, and output gates for protecting and controlling the information flow vector states. In this way, the scale of integration can be changed dynamically at different moments with fixed model parameters, thus avoiding the problem of gradient disappearance or gradient expansion ( Hochreiter et al., 2001 ). The input gate determines how much of the input data of the network at the current moment needs to be saved to the cell state. The forgetting gate determines how much of the cell state needs to be preserved in the current moment from the last moment. The output gate controls how much of the current cell state needs to be output to the current output value. The computation of the LSTM layer can be expressed as follows.
where t i denotes the input gate, ft denotes the forget gate, O t denotes the output gate. C t and C t- 1 denote the state at the current and previous moments, respectively. W is the assigned weight for each layer, x t is the input time step at the current moment, and b is the bias. σ denotes the sigma operation. X denotes the Hadamard product.
The internal structure of the hidden layer of the LSTM is shown in Figure 2 . The forgetting gate f t determines which information coming from the information state h t- 1 at the previous time node needs to be discarded and which needs to be retained. The input information x t from the current moment and h t- 1 from the previous moment are simultaneously fed into the sigmoid activation function, and the output value is the value of the forgetting gate f t . The value range of f t is between (0, 1), and the closer the value is to 1 means that the information passing through the forgetting gate should be retained, and vice versa, it should be discarded. The input gate i t controls which new inputs will be kept in the cell state. The current moment’s input x t and the previous moment’s information state h t-1 are first fed to the sigmoid activation function, which adjusts the value of the input gate i t to a value between (0, 1). Then x i
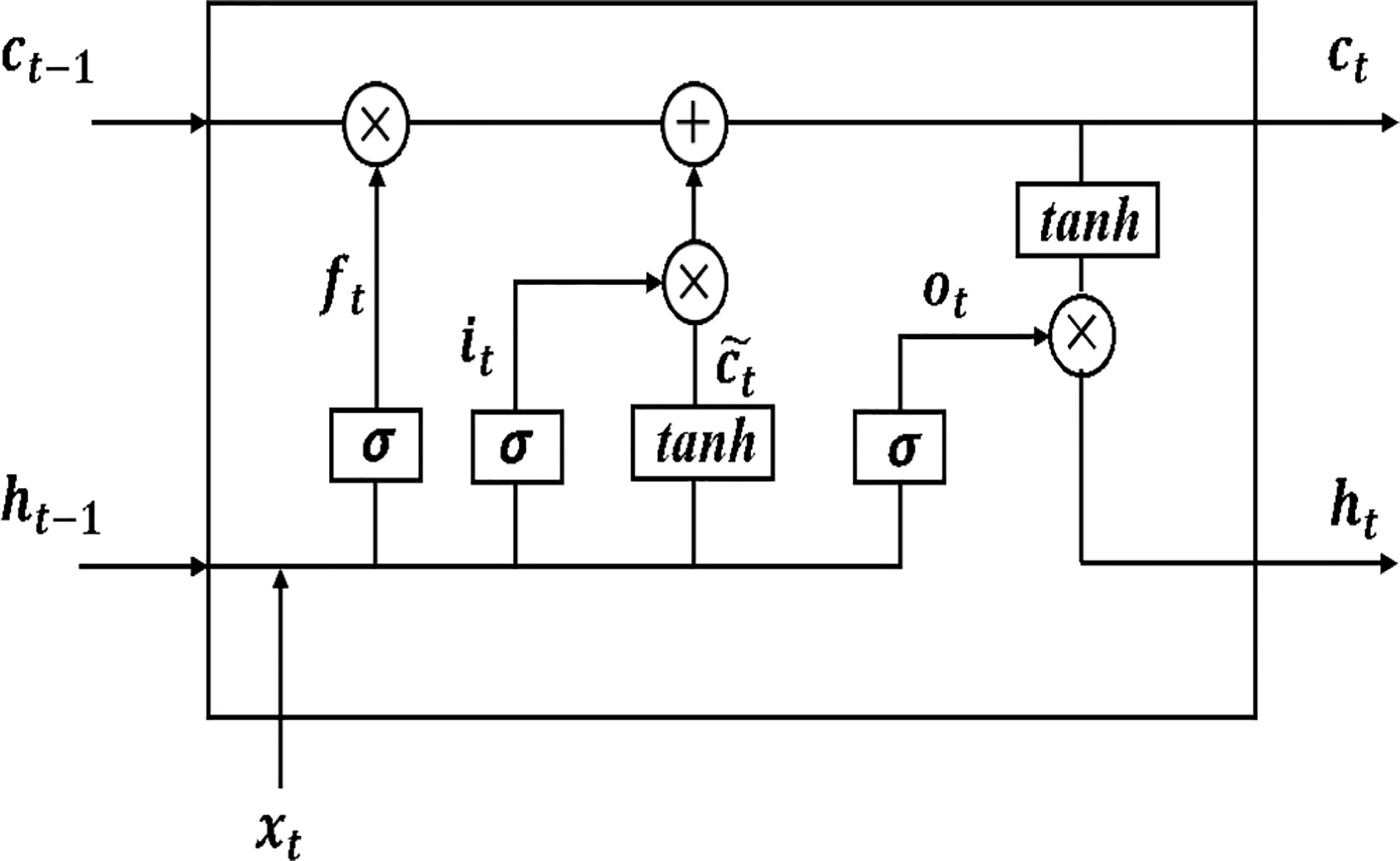
Figure 2 Internal structure of LSTM hidden layer.
and h t-1 are jointly delivered to the tanh function to create a new candidate cell state c t for the current moment, which is followed by the LSTM layer back to update the cell state c t for the current moment. The forgetting gate f t is used to control which information in the previous moment’s cell state c t-1 needs to be discarded, and then the input gate i t is used to determine which information in the current moment’s candidate state c t will be retained in the new cell state, respectively, using the product calculation. Finally, the product of the two is summed to obtain the cell state c t at the current moment. output gate o t controls the output of the current information state, i.e., the information state h t input to the next time node, which is jointly determined by x 1 , h t-1 , and c t .
The limitation of the LSTM application in the ocean domain is that it can only handle time-series data from a single location. It is well known that the ocean is a dynamically changing whole, and different points are temporally and spatially correlated with each other ( Magdalena Matulka and Redondo, 2010 ). Although, researchers can divide the complete ocean into multiple points and use LSTM to process them one by one. However, this approach ignores the regional characteristics of different oceans and the interactions between neighboring points of the same ocean. To address this problem. Shi et al. (2015) improved the LSTM and firstly proposed the Convolutional LSTM Network (ConvLSTM). He and his team use ConvLSTM for rainfall forecasting. They have collected many radar plots which give the distribution of clouds in a given region. Moreover, these maps are changing along the time axis. So with the past timeline and cloud cover maps, it is possible to predict where the clouds should go at future points in time, weather changes, and the chances of future rainfall in an area. Using traditional LSTM models leads to the loss of geolocation information in the cloud cover map, and therefore it is difficult to predict where the clouds will move. The contribution of the original paper is to add the convolution operation that can extract spatial features to the LSTM network that can extract temporal features and propose the architecture of ConvLSTM. ConvLSTM inherits the advantages of traditional LSTM and makes it well suited for Spatio-temporal data due to its internal convolutional structure. The computation of the ConvLSTM layer can be expressed as follows.
where * is convolution operator.
The most important feature of the ConvLSTM algorithm is that it replaces the matrix multiplication in the LSTM with convolution operations. However, its essence is still the same as LSTM, using the previous layer’s output as the input of the next layer. The difference is that with the addition of the convolution operation, the temporal relationships can be obtained, and the spatial features can be extracted like the convolution layer. In this way, Spatio-temporal features can be obtained. Regional wave height forecasting is a typical Spatio-temporal problem. Therefore, the proposed model uses the ConvLSTM algorithm.
3.2 Forecasting method
For the wind wave prediction problem with high spatial and temporal resolution, we would like the proposed model to make longer time predictions. However, if we perform multi-step prediction directly, the error of the results may be unstable. Therefore, the proposed model adopted a different approach from most current forecasting methods that directly establish correlations between specific future moments and historical data. Instead, this study used a more appropriate forecasting strategy to improve the long-term predictive capability of the model. According to previous studies, the Rolling Mechanism (RM) is more suitable for dealing with high-frequency and long-time forecasts ( Akay and Atak, 2007 ; Kumar and Jain, 2010 ).
The main idea of the RM method is to use the obtained forecast data as the latest data and add it to the future forecast. Figure 3 shows the process of forecasting. The small boxes represent the data for each moment. The numbers in the small boxes represent the moments of the data, and the large boxes represent the historical data used for each forecast (time window), which is of length N . Our forecast is a single-step forecast. First, we use the historical data from T-N to the moment T
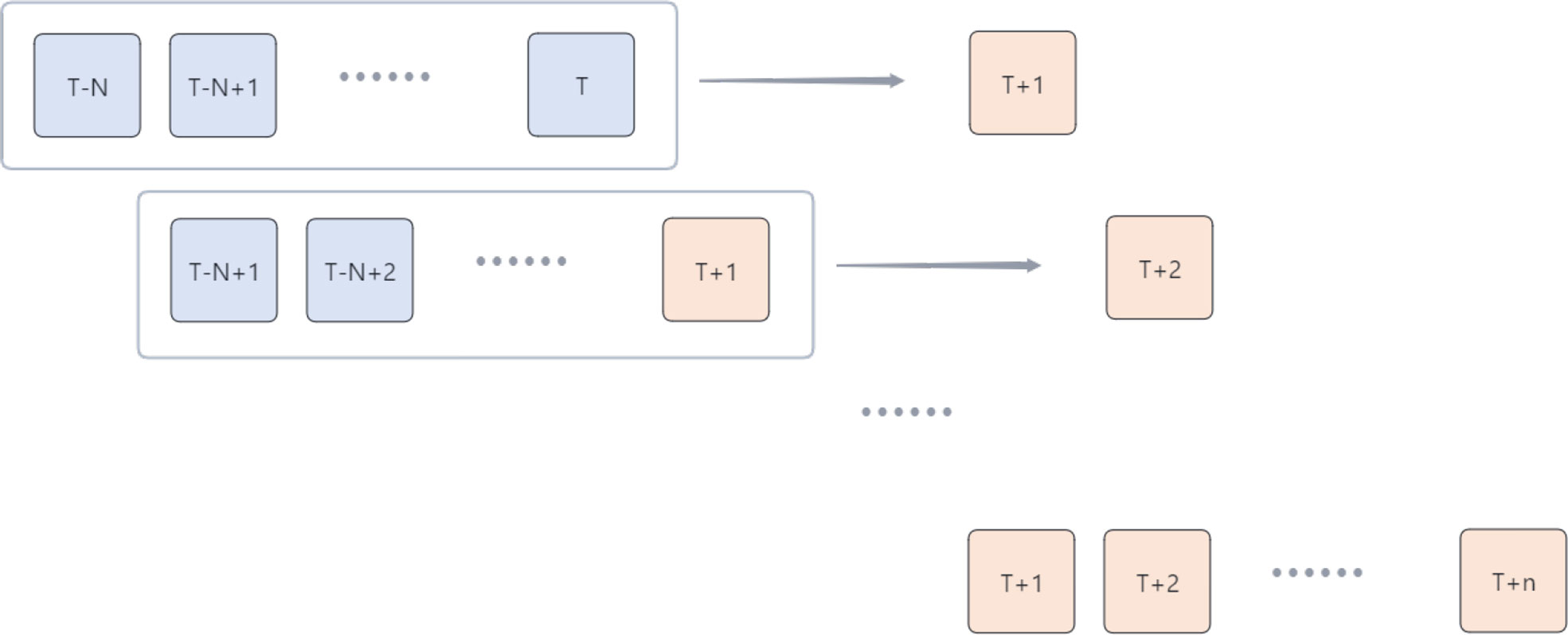
Figure 3 The prediction method integrated with RM.
to forecast the data for the future moment T+1 . Immediately after that, the time window is shifted down by one step. We treat the data just obtained for T+N as known data and use the N data from T-N+1 to T+1 to forecast the data at the moment T+2 , repeating the above process n times. We then get the data from T+1 to T+n moments. In this way, the prediction process of using historical data continuously to predict the next n moments is completed.
It is worth noting that the standard convolution operations in deep learning algorithms such as CNN and ConvLSTM can only act on standard rectangular areas. In our study, the study area is not a pure sea area, but a land-sea combined area, including Hainan Island and parts of the eastern Indo-China Peninsula. Due to the existence of land points, the error calculated during model training will be affected by these land points, thus affecting the effectiveness of the model. This is not what we expected. To solve this problem, we propose the Mask method described below. “Mask” is an idea in deep learning. In simple terms, it is equivalent to masking or selecting some specific elements by putting a mask over the original tensor. This study proposes the Mask method by combining the “Mask” idea.
A brief description of the Mask method is given in Figure 4 . This study generates a matrix of size equal to the input data so that the points in the matrix can correspond to the points in the input data one by one. It will be referred to as the Mask matrix. The antique-white part in Figure 4 represents the land area, and we set the value of the Mask matrix corresponding to this area to 0 and the value of the Mask matrix for the ocean part represented by the blue part to 1. It is worth noting that, in practice, the land-sea distribution of the study area is much more complicated than in Figure 4 . But because of the specificity of the right-angle grid data. We can still build the corresponding Mask matrix according to the idea in Figure 4 . During the model’s training, the model determines the direction of the subsequent gradient descent by calculating the average error between the results and the labels. To implement our Mask method, we rewrite the loss function for network training according to Eqs.11 and 12 during network training.
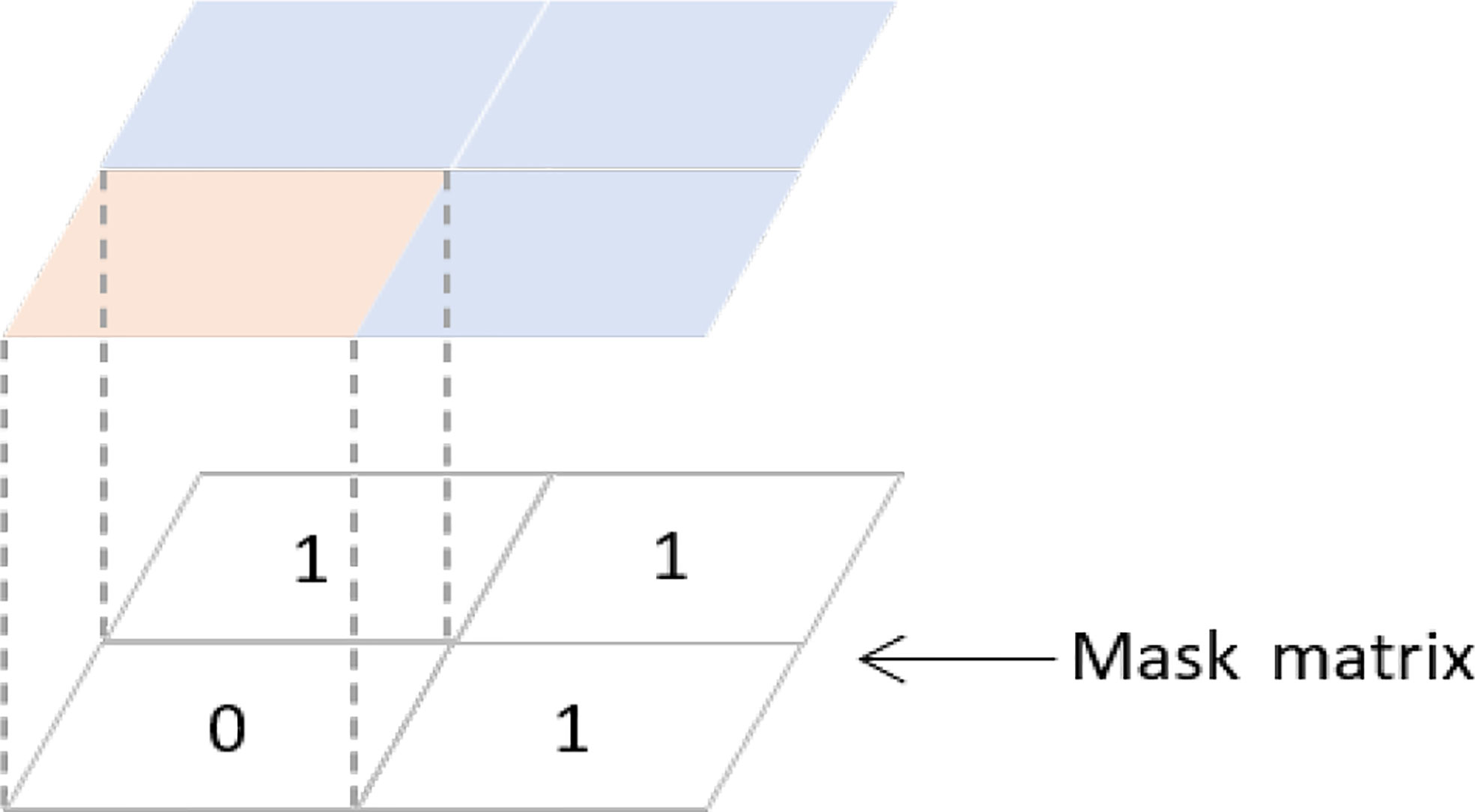
Figure 4 Idea of Mask method. The antique-white areas represent land areas and the blue areas represent ocean areas.
where U denotes the ensemble of points in the study region, Φ land denotes points in the land region, and Φ ocean denotes points in the ocean region. X(t) denotes the output matrix of the prediction at this time. Y t denotes the corresponding factual matrix, and N is the number of points in the ocean area.
In this way, during the training process of the network, the result X (t) of each prediction is subtracted from the control value Y t and then dotted multiplied with the Mask matrix. Since the result is dot multiplied by Mask, and the value of the land part in the Mask matrix is zero, the error of the corresponding land part in the error matrix will also be zero. Therefore, the network only considers the error value of the ocean part in the loss value calculated in each iteration. In this way, the influence of the land region on our experiments is eliminated.
3.3 The proposed model
The wind and wind wave data used in this study have a spatial resolution of 1/40°*1/40° and a temporal resolution of 1 hour. Such high spatial and temporal resolution data means that the degree and speed of sea state variability are much more significant than other slightly lower resolution data. It increases the difficulty of forecasting. As mentioned earlier, there is a strong correlation between wind and wind waves in physical oceanography. If we want to exploit this correlation, we need to have both wind speed and significant wave height as inputs to the model and to have the two dependent on each other. So we need a multi-input network structure to capture this physical correlation and the subtle sea state variations. In this study, we combine the Mask method with ConvLSTM and change the number of input channels of the model to dual channels. It enables the model to meet our needs. The structure of the model proposed in this study is given in Figure 5 .
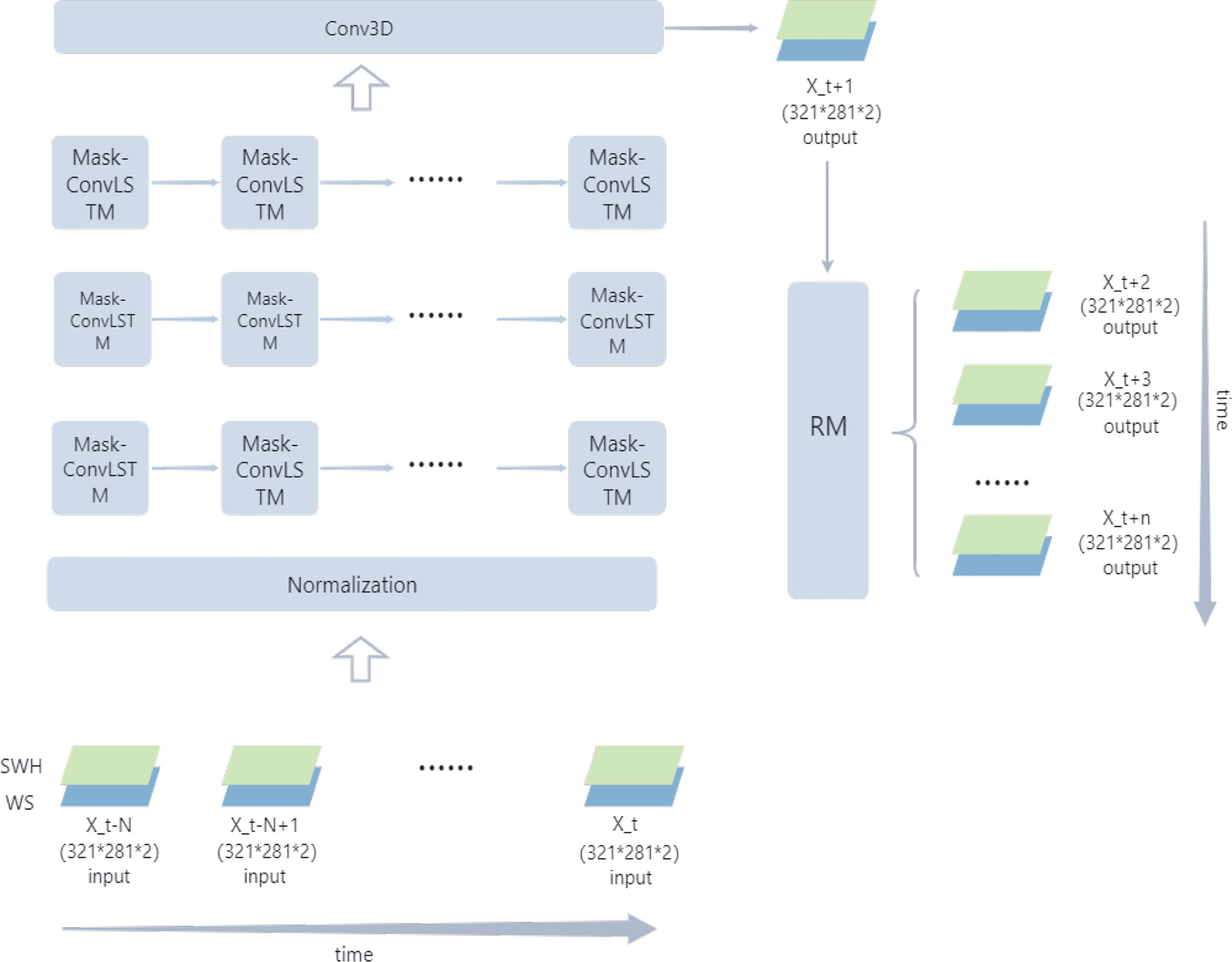
Figure 5 Architecture of the proposed model.
The input data for the model are X t-N to X t . Each input data consists of the wind speed (blue quadrilateral in Figure 5 ) and significant wave height data (green quadrilateral in Figure 5 ) at that moment. The data N is the length of the time window we choose. The size of these input data is (321*281) and after combining them into dual-channel data, each input X has a dimension of (321*281*2). These input data enter the regularization layer for regularization and are then fed to the three Mask-ConvLSTM layers. Between each Mask-ConvLSTM layer, model use relu as the activation function. During the model training process, the network then learns the Spatio-temporal correlation of the input data and the physical correlation between SWH and WS. Then, the size of the output data that we need to obtain is controlled by the convolution layer(Conv3D). In this way, we obtain the predicted data at X t-N moments and then add the obtained predicted data at X t+N moments to the RM module to achieve rolling forecasts.
3.4 Evaluation metrics
In order to evaluate our model reasonably, this study selected a variety of evaluation metrics commonly used to evaluate the significant wave height prediction problem, including Root Mean Square Error (RMSE), Scatter Index (SI), and R Square (R²). In this, the SI can measure the percentage of RMSE relative to the average actual value. However, due to the specificity of the study sea area, we make some modifications to these standard evaluation metrics to match our problem. We combine the indicator RMSE,SI with our Mask method to make it possible to focus only on the error situation in the marine area. It ensures no disturbances from the terrestrial values in the area and that there are no erroneous undercounts due to incorrect, missing point counts. The mathematical equation for these evaluation indicators is as follows:
where X represents the predicted value, Y represents the corresponding value, and N is the number of points in the taken region.
The effect experiments are based on the significant wave height and wind speed data for 2018-01 to 2019-12 mentioned in Section 2. In order to test the performance of the model, this study conducted multiple sets of controlled experiments. The forecast tests were conducted in the validation set that did not participate in the training.
4.1 Performance study
In order to verify the superiority of the proposed model for the high-resolution significant wave height prediction problem, this study compares several published significant wave height prediction methods. The compared methods include the traditional machine learning methods RF ( Callens et al., 2020 ) and SVM ( Berbić et al., 2017 )mentioned in Section 1 and the LSTM ( Fan et al., 2020 ), GRU ( Meng et al., 2021 ) algorithms in deep learning. Five sets of experiments, including the model proposed in this study, used the same training data, and their performance is shown in Figure 6 .
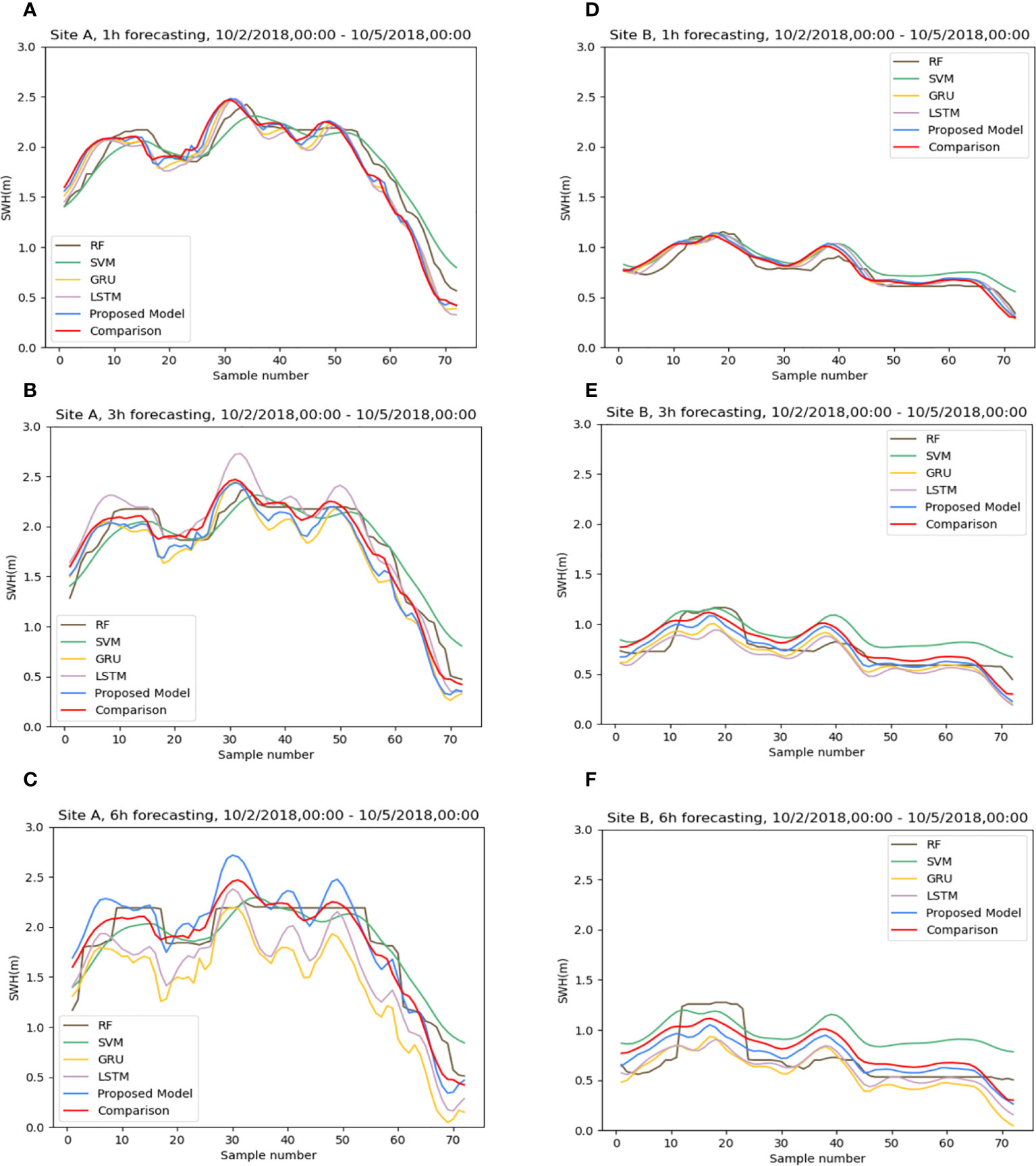
Figure 6 The differences in prediction effects of different algorithms at sites (A, B) : (A, D) 1h, (B, E) 3h, (C, F) 6h. Sample number indicates the time sample number.
Firstly, this study conducted effect experiments for three different forecast lengths of 1 hour, 3 hours, and 6 hours. To more visually show the comparison of the effects between different methods, we chose two sites, siteA(17°N, 110°E) and siteB(19°N,108°E)(shown in Figure 2 ), to conduct our experiments. Figures 6A, D ) show that at a forecast length of 1-h, there is little difference between the predicted and comparison values of the five methods. When the time grows to 3-h, the results of the RF methods show significant differences from the comparison values, especially in the case of low wind waves ( Figure 6E ). Although the SVM algorithm can predict the trend of data variation, the difference in values is significant. When it comes to 6-h, the RF and SVM methods have completely lost their forecasting ability. The effect of the three groups of deep learning algorithms also appears to be very different. Although LSTM, GRU, and ConvLSTM all capture the change in significant wave height, ConvLSTM has more accurate numerical magnitude predictions than the other two.
To avoid the effect of chance, we performed the above experiments for all points in the sea area and averaged the results of the comparisons, which are shown in Figures 7 , 8 . Figure 7A shows the variation of RMSE for the five groups of algorithms. It can be seen that the RMSE of the SVM method has reached about 0.18 at a prediction time of 1 hour. We can also visualize it in Figure 8B . The SVM algorithm lost effectiveness when the predicted large wave height was below 0.5 m. The remaining groups of algorithms do not differ much. The prediction effectiveness of the conventional machine method decays severely with time. Especially for smaller apparent wave heights, both SVM and RF show different degrees of inaccuracy. The RMSE of the SVM algorithm reaches about 0.3 for 3 hours and even 0.7 for 6-hour forecasts, while the RMSE of RF also reaches about 0.6. Two groups of deep learning methods, LSTM and GRU, have acceptable performance in the early stage ( Figure 8A, C . However, the error after 6 hours is also much higher than that of the ConvLSTM method, with the RMSE increasing to more than 0.4 and the SI reaching 0.2. The comparison of the SI of different experimental groups in Figure 7B also shows the superiority of the ConvLSTM algorithm in this study. As we mentioned in Section 3, when dealing with such high spatial and temporal resolution data if only the temporal correlation of one point is considered without considering the spatial relationship of each point. It would be difficult for the model to predict the sea state changes within the sea area accurately. It may be the reason for the poor performance of the LSTM and GRU algorithms in this study.
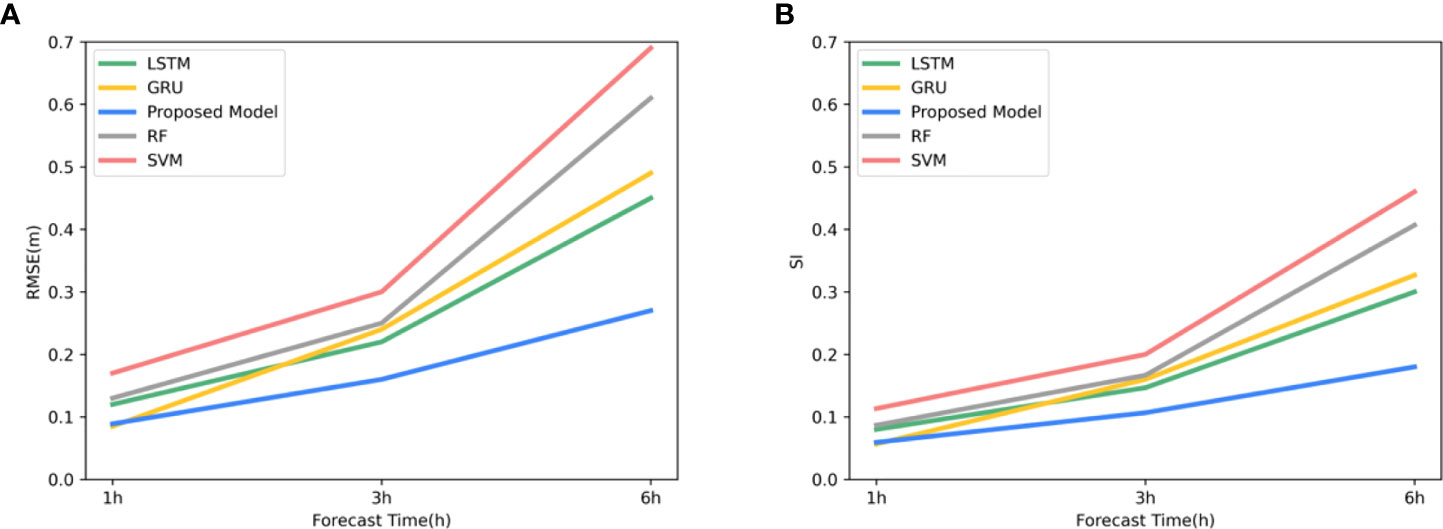
Figure 7 The difference in prediction effect of different algorithms: (A) RMSE of different algorithms varies with the forecast time, and (B) SI of different algorithms varies with the forecast time.
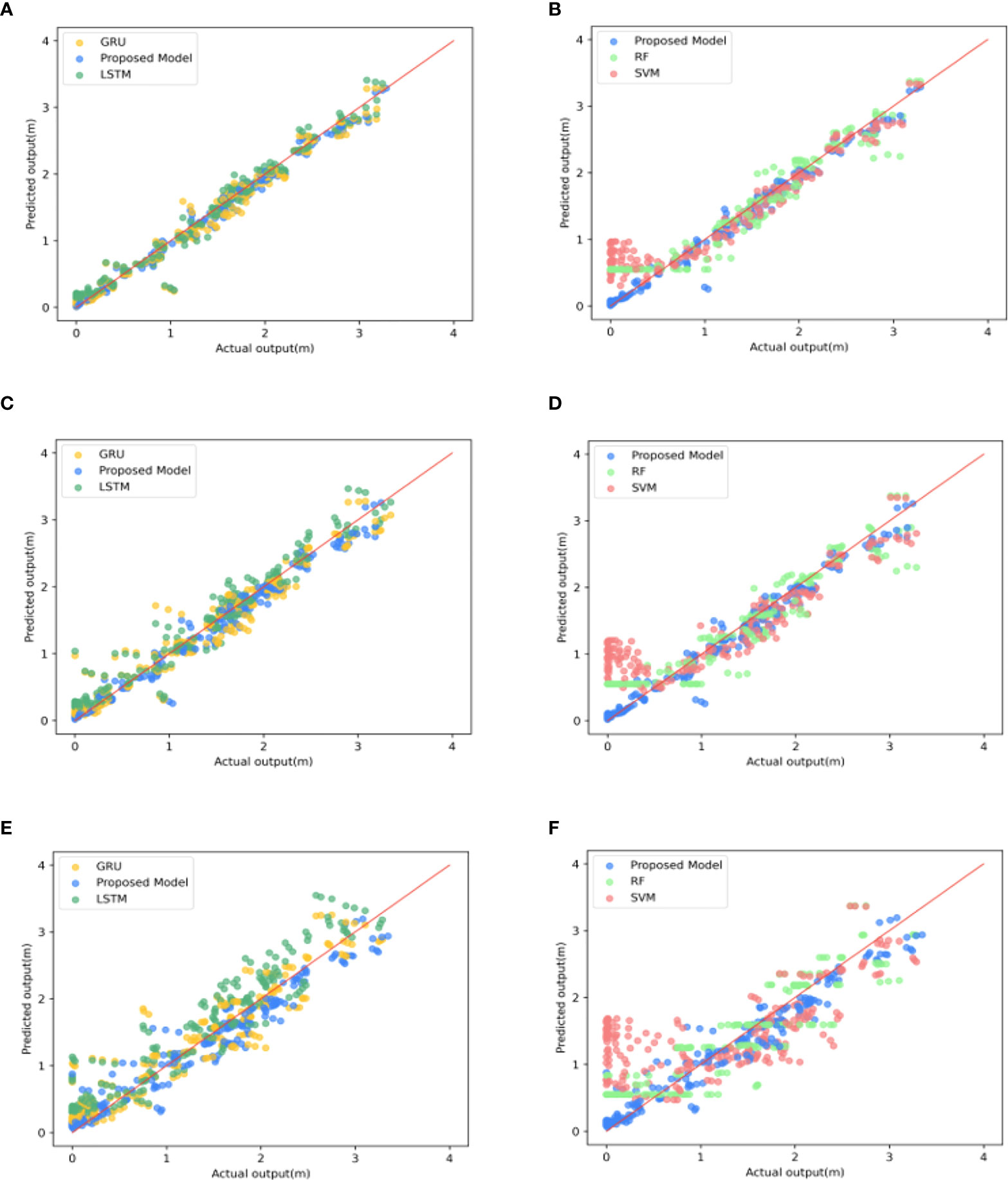
Figure 8 Scatter plots of five groups of algorithms in different forecast times: (A, B) 1 h, (C, D) 3 h, (E, F) 6 h.
4.2 Ablation study
In order to verify the advantages of the wind and wind waves dual-channel compared to the wave-only single channel and the effectiveness of introducing the Mask mechanism, we set up four different sets of experiments. First, a single-channel experiment using only significant wave height data for training and no Mask method (Single-channel). Second, single-channel experiments use only significant wave height data for training but with the Mask method (Single-channel & Mask). Third, experiments using significant wave height and wind speed data for training but without using the Mask method (Dual-channel). Finally, experiments using the significant wave height and wind speed for training with the Mask method (Dual-channel & Mask). It is worth mentioning that the results obtained by the dual-channel network during the prediction process are also dual-channel, containing both the prediction results of the significant wave height and the wind speed. This study aim to study the network’s prediction of the significant wave height. Therefore, we take only the significant wave height prediction results from the dual-channel network forecast results when comparing different groups of experiments.
To ensure the validity of these four sets of experiments. The hyperparameters of the four sets of experiments will be set to the same set of values. We randomly selected a period of history and made a 6-hour forecast downward using each of the four models. Figure 9 shows the results of our experiments. It can be seen that the error of the dual-channel network is significantly smaller than that of the single-channel network when only the number of channels is considered, and this situation persists with increasing forecast time. It proves the advantage of the dual-channel network. With the introduction of wind speed, the network takes advantage of the physical correlation between the two to improve our forecasting results for the significant wave height. Similarly, we can see that after the Mask method is used. When the channels are the same, the Mask-RMSE in the two groups of experiments using the Mask method is smaller than in the other two groups of experiments without the Mask method. However, we noticed a particular case. Before 3-h, the error of the single-channel network with the Mask method is smaller than that of the dual-channel network without the Mask method. It may be because the Mask mechanism can dominate the error situation brought by the forecast in the short term. However, this slowly disappears as the forecast time goes on, and both sets of dual-channel experiments slowly outperform the two sets of single-channel experiments. The above findings demonstrate the superiority of the wind and wind waves dual-channel network compared to the wave single-channel network and the effectiveness of the Mask method in this experiment.
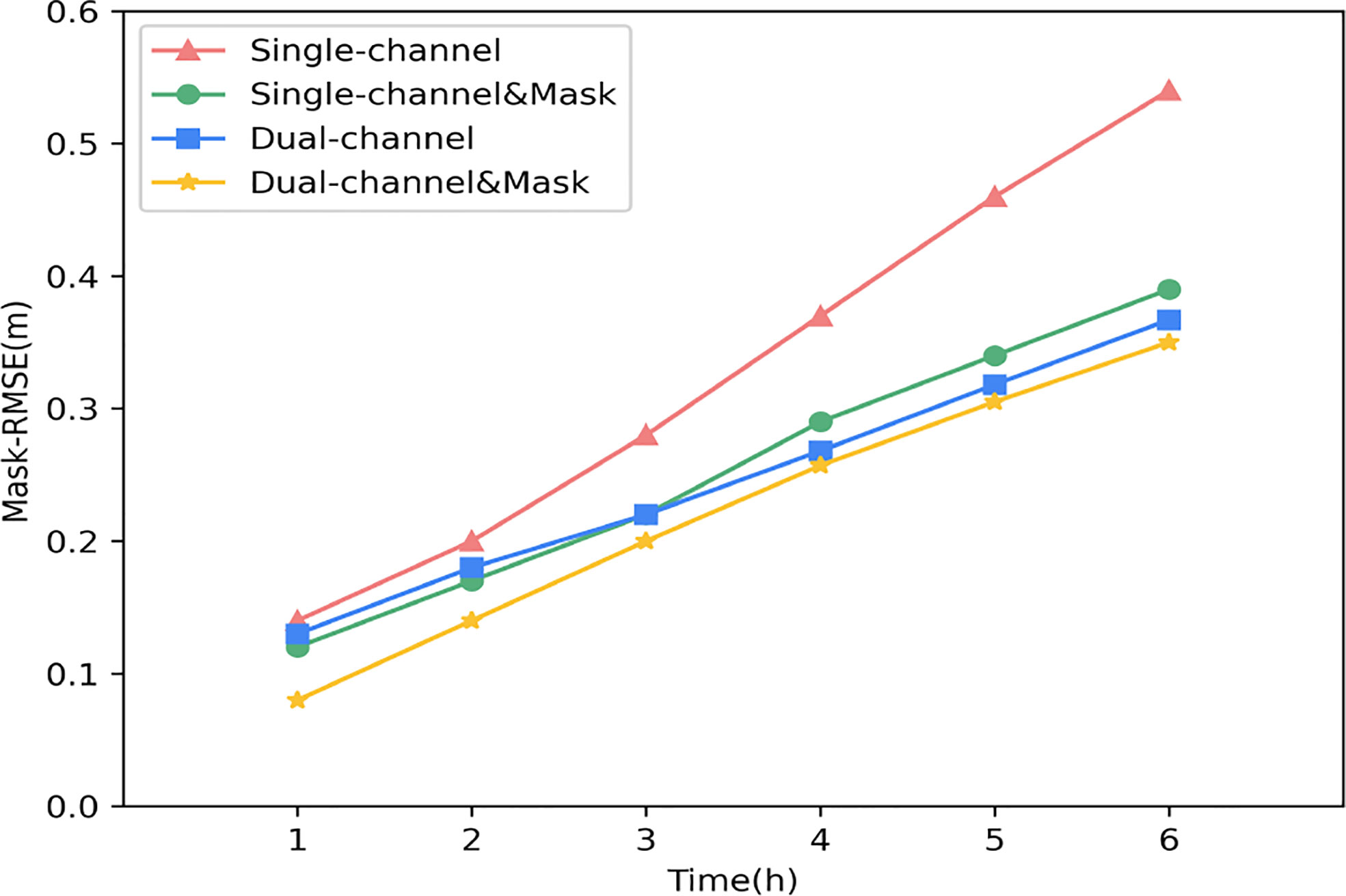
Figure 9 Mask-RMSE of the four groups varied with the forecast times.
4.3 Conventional forecast
After completing the training process of the model, we first explore the performance of the proposed model on intermediate time scales. As input data, we use 6 hours of data from 00:00-12:00 on August 28, 2019. To more visually demonstrate the ability of the proposed model to predict the high-resolution significant wave height of the study sea, we plot the results. Results and error statistics are shown in Figure 10 and Table 1 .
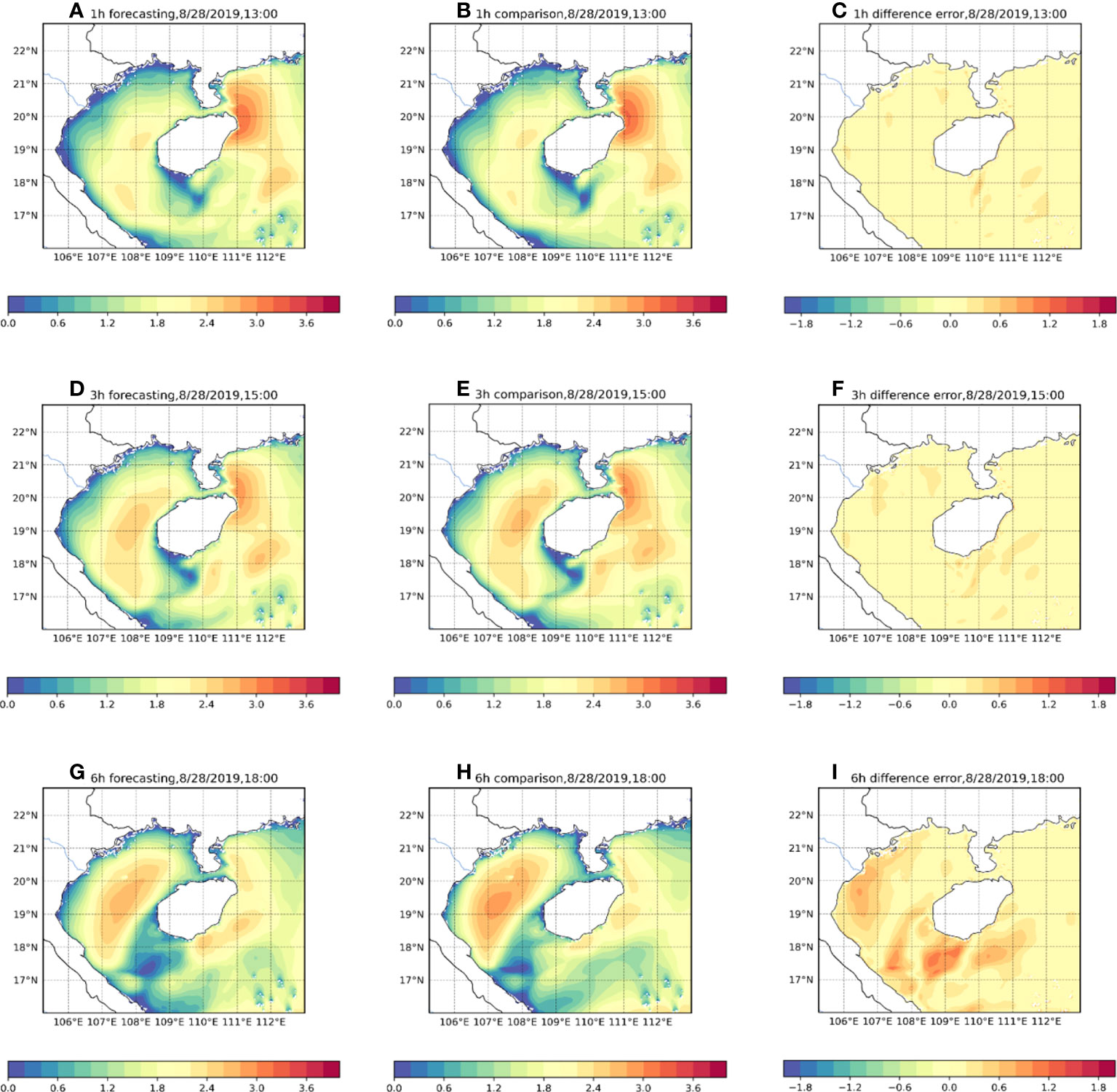
Figure 10 Comparison of forecast effect of the model under different forecast times. (A, D, G) are the predicted significant wave height effect diagrams for 1h, 3h, and 6h, respectively. (B, E, H) is the significant wave height diagram of the numerical model at the corresponding time. (C, F, I) is the difference error between forecasting and comparison.
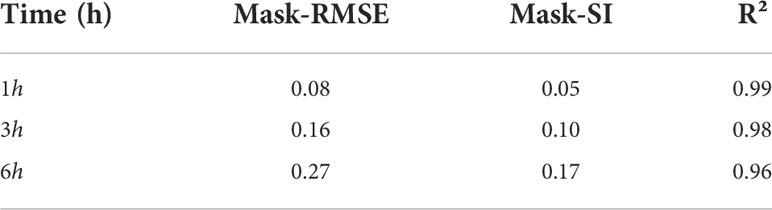
Table 1 The evaluation metrics of the model at different forecast time.
In Figure 10 , the three subplots (a), (d) and (g) on the left side represent the significant wave height forecasts for 1,3,6-h, respectively. The numerical model data at the corresponding time are shown on the right side. Their evaluation indicators are shown in Table 1 . We can see from Figures 10A , B that our model accurately captures the distribution of the significant wave height at 1-h with a Mask-RMSE of only 0.08 and accurately predicts the higher wind waves in the northwestern part of Hainan Island. After increasing the time to 3-h, the proposed model still has high accuracy and low error value, and the R² value can be maintained at 0.98. It also has a good prediction ability for distributing significant wave height and wave height size in the region. However, numerically, the predicted values for the western part of Hainan Island in Figure 10D are smaller than the corresponding values. When the time window comes to 6-h, although our model can still capture the significant wave height distribution in the sea, there is a significant difference in the values( Figures 10G, H ). Mask-RMSE increases to 0.27. However, this value is perfectly acceptable for such high spatial and temporal resolution data. Overall, the forecasting effect of our model is excellent.
4.4 Analysis of error sources and treatment
The model we use is a dual-channel network. In the prediction process, both SWH and WS channels generate errors. Because the network considers the characteristic correspondence between wind and wind waves, if the error in one channel is too large, it will also decrease the prediction of the other channel. In order to study the variation of the error of the two channels with the forecast time during the forecast, we extended the forecast time to 12-h in Section.4.3 and analyzed the error sources.
For the experimental results in Section.4.3, we analyze the error variance of the two channels in the prediction process separately. The yellow line in Figure 11 shows the trend of the prediction error of WS over time, and the blue line shows the trend of the error of SWH. It can be seen that the Mask-RMSE of both channels increases with the prediction time due to the rolling mechanism. However, on the way up, the blue line shows a steady upward trend over time, while the yellow line shows a strong upward trend, especially after 6 hours. It shows that the predictive validity of the WS channel drops significantly after 6 hours. The correlation between the two channels is considered in the model prediction process. The sharp increase in the error of the WS channel will directly lead to an increase in the error of the SWH channel, increasing the total error of the prediction results. We pioneered a new mechanism called Replace and added the Replace mechanism to the RM process to solve this problem.

Figure 11 The Mask-RMSE of significant wave height and wind speed changes with increasing forecast time.
The core idea of the Replace mechanism is to replace the predicted wind speed values of the wind speed channel in the network with the wind speed of the numerical model. As shown in Figure 12 , the model is run to get the forecast output X t+1 , and the RM mechanism uses the obtained X t+1 to continue the rolling forecast to get the output results for the next n-1 moments. The black dashed box in Figure 12 explains the specific steps of the Replace mechanism. As we mentioned before, the forecast results of each step of the model include forecast SWH and forecast WS. However, the error of WS increases sharply during the rolling forecast process ( Figure 11 ), leading to an increase in the error of SWH associated with it. Suppose we can solve the problem of a sharp increase of WS in this process. Then the forecast results will be improved. To solve this problem, we perform the following operation for each model and RM mechanism forecast result X t+m : replace the WS obtained from the network forecast with the WS of the numerical model to form a new X t+m consisting of the numerical model WS and the network forecast SWH (red dashed box in Figure 12 ). This data is used to replace the original X t+m for RM processing.
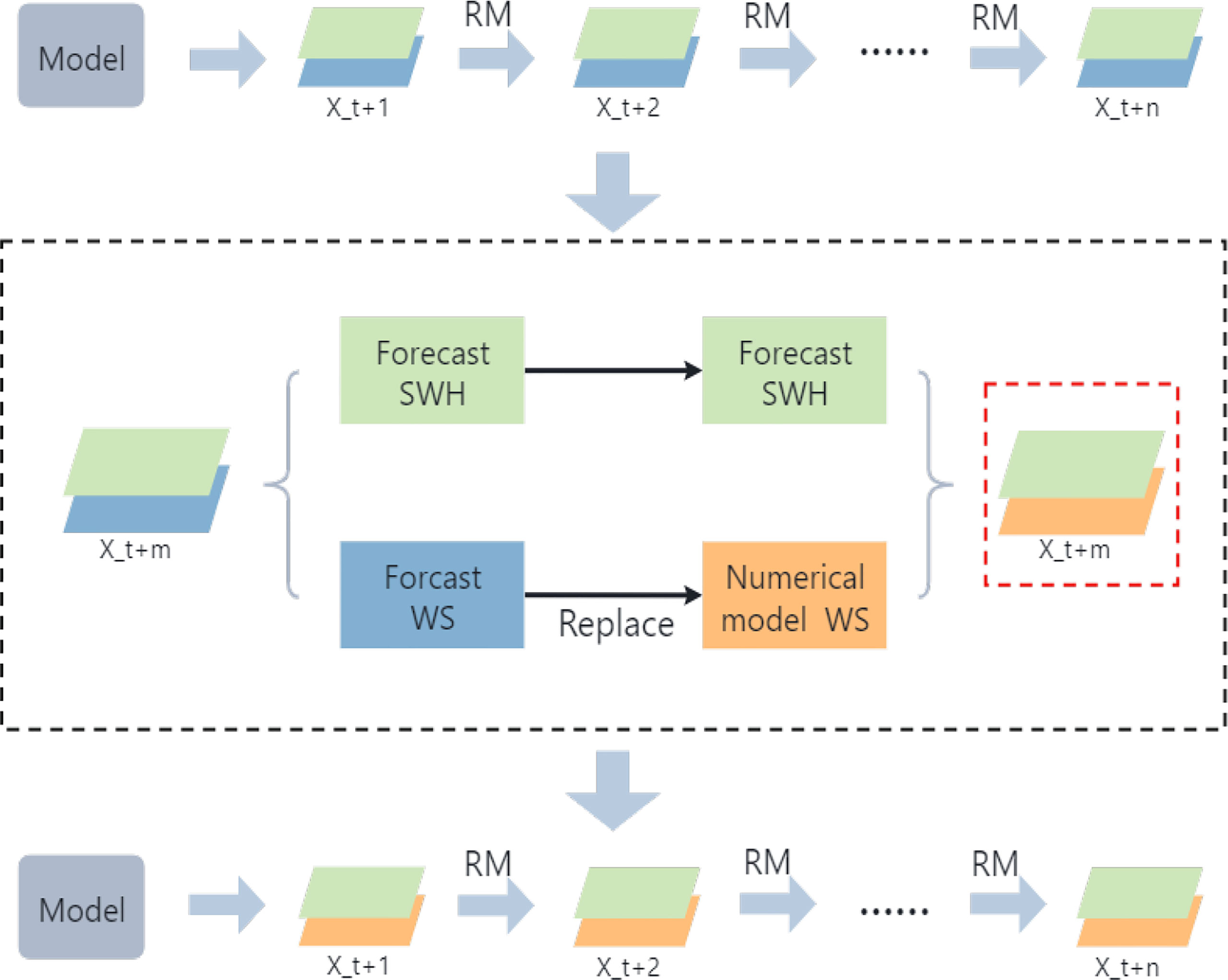
Figure 12 The idea of Replace Mechanism. The black dotted box represents the core steps of the RM mechanism, and the red box represents the data obtained through the RM mechanism.
To verify the effectiveness of the Replace mechanism, we conducted a set of controlled experiments. The error profiles of SWH before and after adopting the Replace mechanism are shown in Figure 13 . The yellow line is the error plot of SWH after adopting the Replace mechanism, and the blue line is the forecast error without the Replace mechanism. It can be seen that the adoption of the Replace mechanism significantly reduces the overall forecast error, especially in the medium and long time scales. Compared to the previous one, the Mask-RMSE is even reduced by up to 50%.
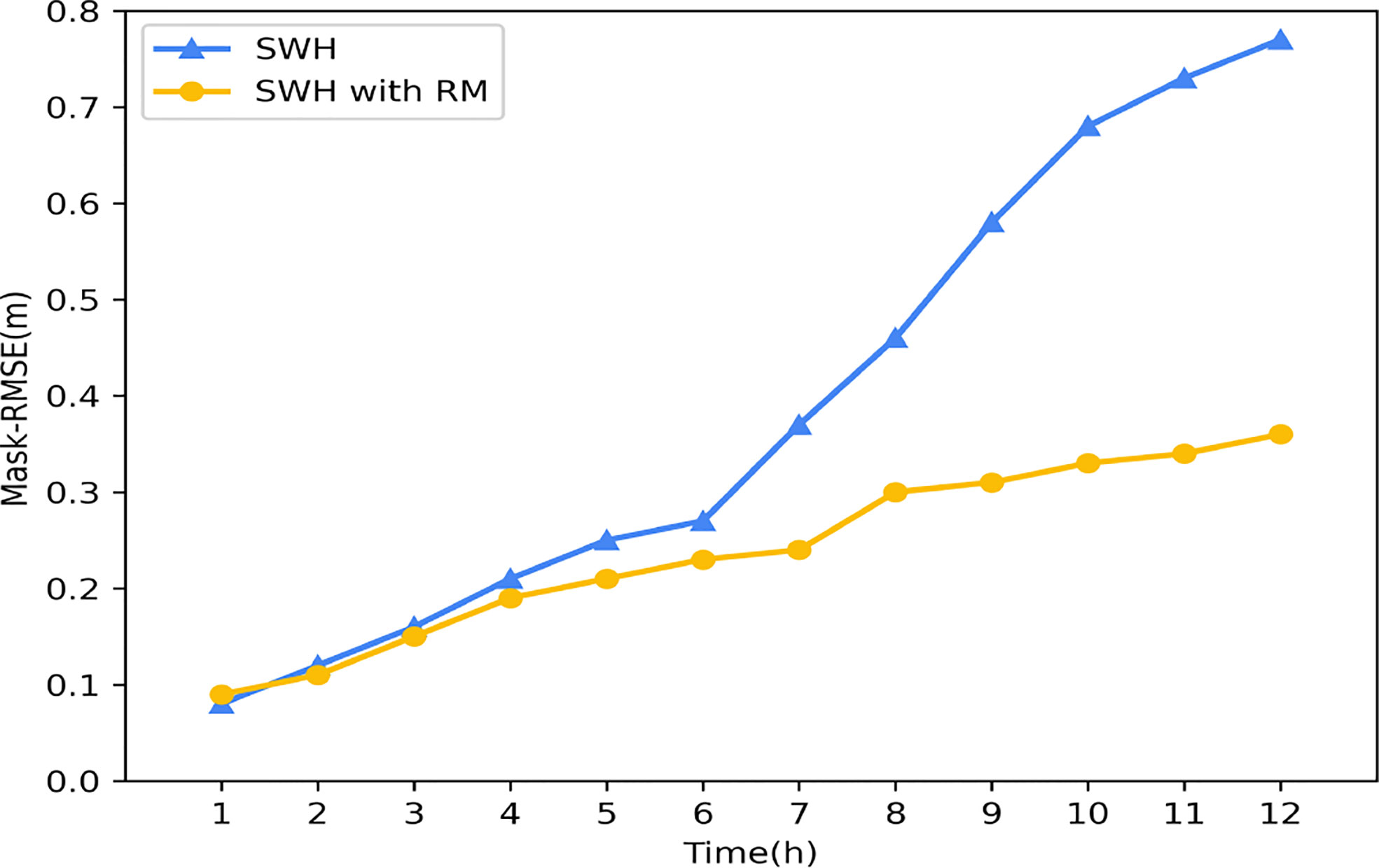
Figure 13 Comparison diagram of Mask-RMSE changes of the model before and after using the RM mechanism.
4.5 Long time scale forecasting
In previous deep learning wave height forecasting studies. Within the margin of error, the effective forecast duration obtained using hourly data was typically limited to 6-12 hours. If the data resolution is increased, this time will be further shortened. To investigate whether the proposed model can make predictions on long time series after using the Replace mechanism, we set up two sets of experiments. One group had Replace mechanism, and the other group had no Replace mechanism. We adjusted the timestep to 12 and retrained the model using previous data from each set of experiments. The results of the two experiments are shown in Figure 14 , where (a) is the numerical model data, (b) is the prediction result of the model with the Replace mechanism, and (c) is the prediction result of the model without the Replace mechanism.
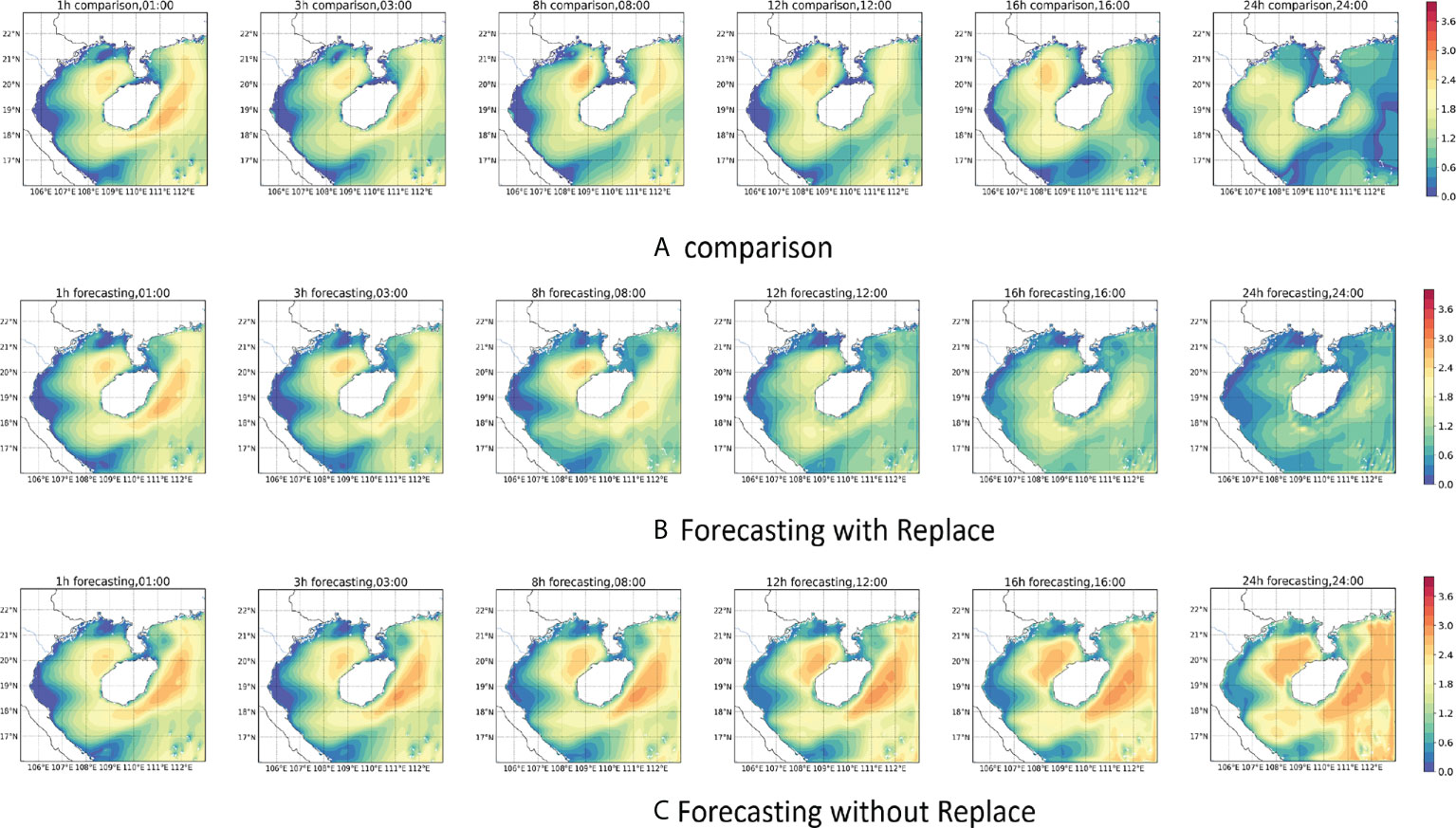
Figure 14 24-hour forecast results of significant wave height. (A) is the significant wave height diagram of the numerical model. (B) is the predicted significant wave height diagram after Replace mechanism is adopted in the model. (C) is the predicted significant wave height diagram without Replace mechanism.
We predicted the significant wave height for the next 24-hour using the data from 12:00-24:00 on August 2, 2019. The variation of each indicator with time is shown in Table 2 . It can be seen that the two sets of experiments still maintain good performance at 1-hour and 3-hour ( Figures 14B, C ). The significant wave height and the distribution of wind waves in the sea still have a good forecasting effect. When the time reached 8-hour, the effects of the two groups of experiments began to show more apparent differences. The network without the Replace mechanism has a higher prediction of the significant wave height in the eastern part of Hainan Island. In contrast, the network with the Replace mechanism can still predict the change of the significant wave height in the sea more accurately, but the distribution has slightly deviated. When we increase the prediction scale to 12-hour, it can be seen that the model without the Replace mechanism has a significant prediction error for the change of significant wave height in the sea, and R² drops to 0.75, basically losing the accuracy of the forecast. However, the model with the replacement mechanism still has good forecasting ability, with a Mask-RMSE of 0.29. Although there are some regions where the significant wave height values are under-predicted, the overall distribution can still be predicted more accurately. When the prediction scale is expanded to 16-hour, the model without the replacement mechanism has lost its predictive power, with R² dropping to 0.62. The model with the replacement mechanism also shows a decrease in accuracy. Although the high wind and wave area near Hainan Island can be predicted, the magnitude of the significant wave height obtained from the prediction has been significantly different, with R-squared dropping to 0.78. When the time reaches 24-hour, the model without the replacement mechanism already shows confusion in the forecasts. The forecasts obtained from the predictions have little to no relationship with the contrasting values ( Figure 14A ). Although the model with the replacement mechanism can predict the distribution of significant wave heights in the sea area, the prediction of the magnitude of significant wave heights in the whole sea area also shows a more noticeable difference, and the R-squared drops to 0.69. Through the above experiments, we can see that: the Replace mechanism is very effective in improving the forecasts of the dual-channel network. In particular, it can effectively reduce the forecast error in a more extended time range. With the Replace mechanism, the long-time forecasting capability of the network is substantially improved, and the significant wave height at high spatial and temporal frequencies within 24 hours can be predicted more accurately.
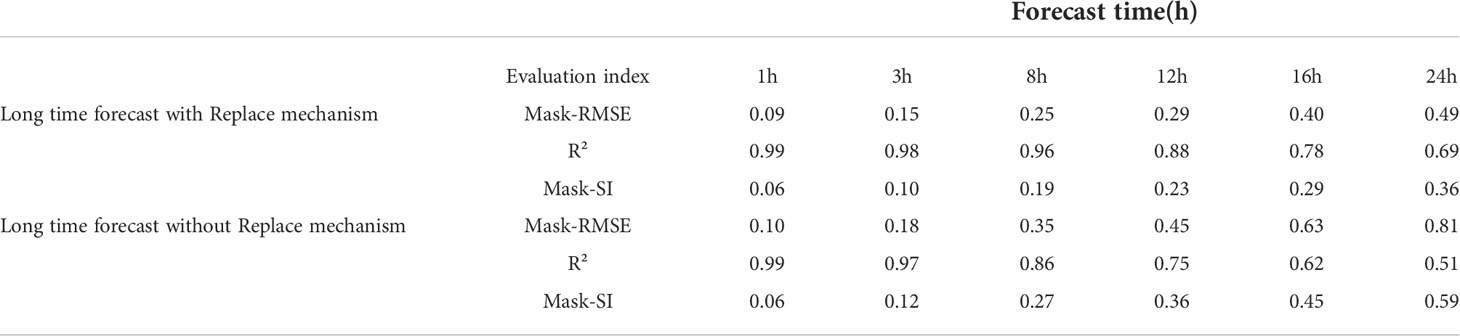
Table 2 The Evaluation index of the two model at different forecast time.
In order to verify the stability of the model’s forecasting ability, this study selected the data of SiteA and SiteB for the whole of October 2018, with a total of 744 samples. Moreover, forecast tests with different lengths were conducted using these data, and the results are shown in Figure 15 . The red line in Figure 15 represent the comparison values. The yellow line represents the ERA5 reanalysis data. The blue line represents the forecast values of the model. It can be seen that the model has an excellent forecasting effect within 6-h. Both the magnitude and the trend of the values differ very little from the comparison values. When the time comes to 12-h, the model can predict the trend of the significant wave height. However, the values are not stable. When the time comes to 24-h, The model can predict the primary trend, but the values are much different in some cases. It may be because our forecasts are regional, focusing on the trend of the significant wave height within the whole region rather than the significant wave height at a single point. In addition, the insufficient amount of data in this study, which only used two years of data, might be another reason.
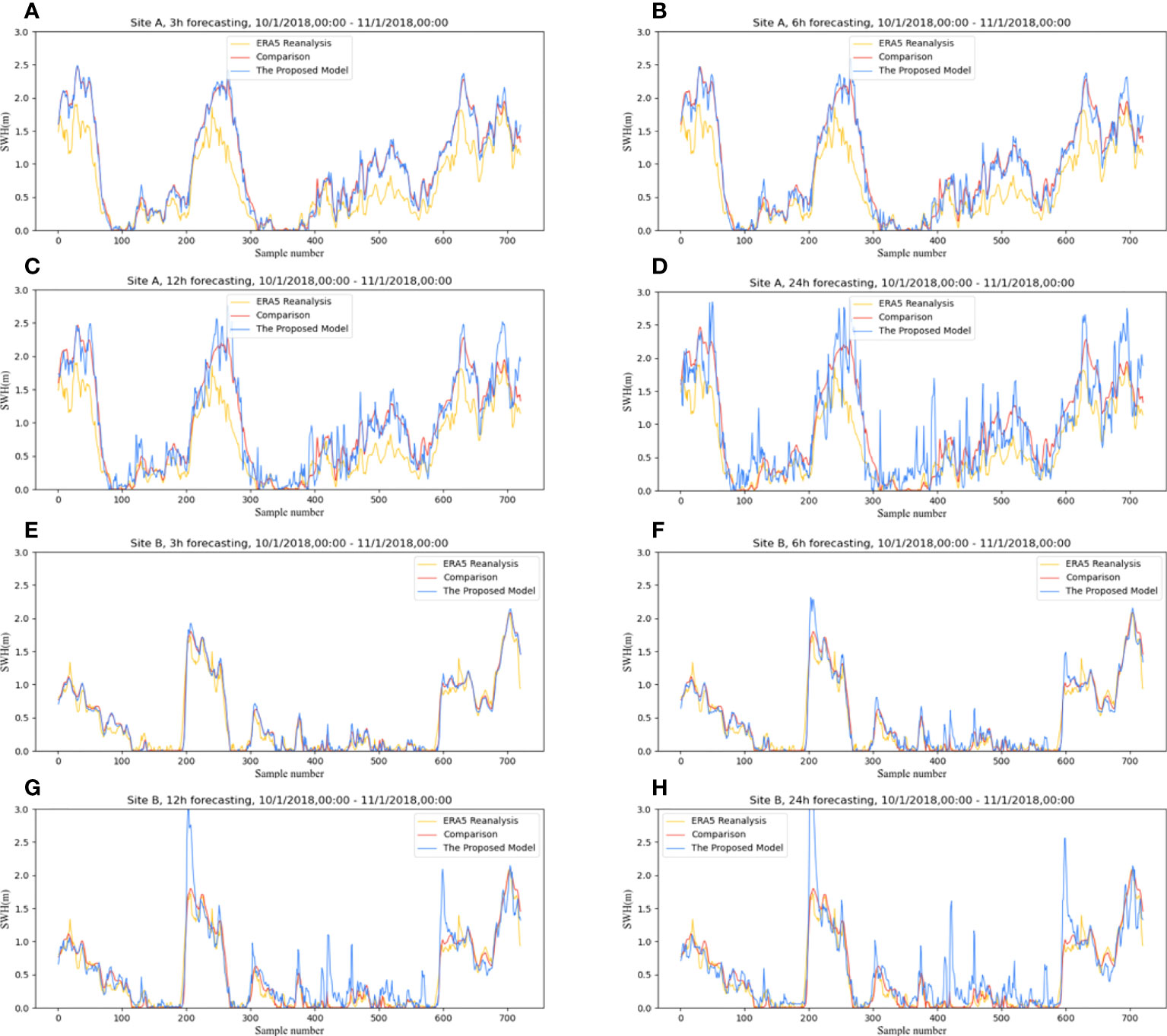
Figure 15 24-hour prediction results of significant wave heights for SiteA and SiteB. (A–D) respectively represent the 3h, 6h, 12h and 24h forecast results of SiteA, and (E–H) respectively represent the 3h, 6h, 12h and 24h forecast results of SiteB.
5 Conclusion and discussion
The main work of this research is to combine the laws of physical oceanography and use the deep learning method to predict the significant wave height of the entire Beibu Gulf with high spatial and temporal resolution. This study uses a modified ConvLSTM network to explore the Spatio-temporal correlation of historical data and the physical correlation between different marine elements. By comparing with other methods, we show the advantages of the proposed model in dealing with Spatio-temporal data. By introducing a two-dimensional wind speed field, the significant wave height prediction capability of the network is greatly improved. At the same time, this study proposes a Mask method to solve the problem of wave height prediction in sea areas with land areas. This study uses high-resolution wind speed and significant wave height data from 2018 to 2019 to conduct forecast experiments on different forecast time scales. The validity of the model is demonstrated by comparison with the numerical model. The R-squared for the 6-hour forecast reached 0.96. We analyze the error generation during the prediction process and propose an alternative mechanism to alleviate the error diffusion problem of the proposed model and prolong the effective prediction time of the network. The effective forecast time reaches 24 hours. Most importantly, the proposed model has practical application value. We know that numerical models predict waves with the wind as input. The method of this study can also utilize these wind data for wave forecasting and compare the results with short-term numerical models. It will significantly reduce the cost issues associated with numerical models. Typically, the time cost for a numerical model run to complete a prediction is over several hours and requires supercomputer support. In contrast, the model proposed in this study can be run on an ordinary PC, and the time needed for prediction is only a few seconds. It is worth pointing out that the present study was conducted for wind waves. The method proposed in this study may not be effective for a region where the wavesare mainly swell-dominated.
The proposed method also has specific problems. The first is that the RM mechanism leads to the accumulation of errors in the prediction process, and our proposed replacement mechanism can alleviate this problem to some extent. However, it cannot fundamentally solve this problem. Secondly, although the proposed model can learn some physical laws of wind speed and significant wave height, it does not really incorporate the dynamic process of the ocean in a sense. How to combine deep learning with the dynamic processes of the ocean is a problem we need to solve in the future. Thirdly, this study is only for the significant wave height, but there are other elements such as wave direction and period. If possible, we will follow up on these elements as well. Finally, we will open up the high spatial and temporal resolution data used in this work. These data will beof great help for subsequent studies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://doi.org/10.5281/zenodo.6402321 .
Author contributions
TS provided overall ideas, writing review and editing, supervision, project administration, and funding acquisition. RH provided concepts, methodology, software, manuscript writing, preparation and research. FM provides formal analysis, data processing, software, and supervision. JW provided data collation and formal analysis. WW provided data management, software. SP provided methodology, method evaluation, data management, project management, and funding acquisition. All authors contributed to the article and approved the submitted version.
Innovation fund project for graduate students of China University of Petroleum (East China) (No. 22CX04008A). Project Supported by Key Laboratory of Environmental Change and Natural Disaster of Ministry of Education, Beijing Normal University (Project No. 2022-KF-08).
Acknowledgments
Over the course of my researching and writing this paper, I would like to express my thanks to all those who have helped me. A special acknowledgement should be shown to SP, from whose lectures I benefited greatly, I am particularly indebted to SP who gave me kind encouragement and useful instruction all through my writing. Moreover, I wish to extend my thanks to the library and the electronic reading room for their providing much useful information for my thesis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akay D., Atak M. (2007). Grey prediction with rolling mechanism for electricity demand forecasting of turkey. Energy 32, 1670–1675. doi: 10.1016/j.energy.2006.11.014
CrossRef Full Text | Google Scholar
Barnett T. P. (1968). On the generation, dissipation, and prediction of ocean wind waves. J. Geophysical Res. 73, 513–529. doi: 10.1029/jb073i002p00513
Barnett T., Kenyon K. (1975). Recent advances in the study of wind waves. Rep. Prog. Phys. 38, 667. doi: 10.1088/0034-4885/38/6/001
Berbić J., Ocvirk E., Carević D., Lončar G. (2017). Application of neural networks and support vector machine for significant wave height prediction. Oceanologia 59, 331–349. doi: 10.1016/j.oceano.2017.03.007
Bolton T., Zanna L. (2019). Applications of deep learning to ocean data inference and subgrid parameterization. J. Adv. Modeling Earth Syst. 11, 376–399. doi: 10.1029/2018ms001472
Booij N., Ris R. C., Holthuijsen L. H. (1999). A third-generation wave model for coastal regions: 1. model description and validation. J. geophysical research: Oceans 104, 7649–7666. doi: 10.1029/98jc02622
Callens A., Morichon D., Abadie S., Delpey M., Liquet B. (2020). Using random forest and gradient boosting trees to improve wave forecast at a specific location. Appl. Ocean Res. 104, 102339. doi: 10.1016/j.apor.2020.102339
Cooke N., Li T., Anderson J. A. (2011). The tongking gulf through history (Philadelphia, United States: University of Pennsylvania Press).
Google Scholar
Fan S., Xiao N., Dong S. (2020). A novel model to predict significant wave height based on long short-term memory network. Ocean Eng. 205, 107298. doi: 10.1016/j.oceaneng.2020.107298
Fu Y., Zhou X., Sun W., Tang Q. (2019). Hybrid model combining empirical mode decomposition, singular spectrum analysis, and least squares for satellite-derived sea-level anomaly prediction. Int. J. Remote Sens. 40, 7817–7829. doi: 10.1080/01431161.2019.1606959
Gao J., Chen B., Shi M. (2015). Summer circulation structure and formation mechanism in the beibu gulf. Sci. China Earth Sci. 58, 286–299. doi: 10.1007/s11430-014-4916-2
Gavrikov A., Krinitsky M., Grigorieva V. (2016). Modification of globwave satellite altimetry database for sea wave field diagnostics. Oceanology 56, 301–306. doi: 10.1134/s0001437016020065
Graves A., Schmidhuber J. (2005). Framewise phoneme classification with bidirectional lstm and other neural network architectures. Neural Networks 18, 602–610. doi: 10.1016/j.neunet.2005.06.042
PubMed Abstract | CrossRef Full Text | Google Scholar
Hasselmann K. (1968). “Weak-interaction theory of ocean waves,” in Basic developments in fluid dynamics (Cambridge, United States: Academic Press), 117–182. doi: 10.1016/b978-0-12-395520-3.50008-6
Hochreiter S., Bengio Y., Frasconi P., Schmidhuber J., et al. (2001). Gradient flow in recurrent nets: the difficulty of learning long-term dependencies . IEEE 2001, 237–243. doi: 10.1109/9780470544037.ch14. Dataset.
Hochreiter S., Schmidhuber J. (1997). Long short-term memory. Neural Comput. 9, 1735–1780. doi: 10.1162/neco.1997.9.8.1735
James S. C., Zhang Y., O’Donncha F. (2018). A machine learning framework to forecast wave conditions. Coast. Eng. 137, 1–10. doi: 10.1016/j.coastaleng.2018.03.004
Jönsson A., Broman B., Rahm L. (2003). Variations in the baltic sea wave fields. Ocean Eng. 30, 107–126. doi: 10.1016/s0029-8018(01)00103-2
Kamranzad B., Etemad-Shahidi A., Kazeminezhad M. (2011). Wave height forecasting in dayyer, the persian gulf. Ocean Eng. 38, 248–255. doi: 10.1016/j.oceaneng.2010.10.004
Kirby J. T. (1985). “Water wave propagation over uneven bottoms,” in Tech. rep. (Florida, United States: FLORIDA UNIV GAINESVILLE DEPT OF COASTAL AND OCEANOGRAPHIC ENGINEERING).
Koongolla J. B., Lin L., Pan Y.-F., Yang C.-P., Sun D.-R., Liu S., et al. (2020). Occurrence of microplastics in gastrointestinal tracts and gills of fish from beibu gulf, south china sea. Environ. pollut. 258, 113734. doi: 10.1016/j.envpol.2019.113734
Kumar U., Jain V. K. (2010). Time series models (grey-markov, grey model with rolling mechanism and singular spectrum analysis) to forecast energy consumption in india. Energy 35, 1709–1716. doi: 10.1016/j.energy.2009.12.021
Li S., Li Y., Peng S., Qi Z. (2021). The inter-annual variations of the significant wave height in the western north pacific and south china sea region. Climate Dynamics 56, 3065–3080. doi: 10.1007/s00382-021-05636-9
Longuet-Higgins M. (1963). The generation of capillary waves by steep gravity waves. J. Fluid Mechanics 16, 138–159. doi: 10.1017/s0022112063000641
Magdalena Matulka A., Redondo J. M. (2010). Mixing and vorticity structure in stratified oceans. in. EGU Gen. Assembly Conf. Abstracts. , 424. Available at: https://ui.adsabs.harvard.edu/abs/2010EGUGA..1215573M
Meng F., Song T., Xu D., Xie P., Li Y. (2021). Forecasting tropical cyclones wave height using bidirectional gated recurrent unit. Ocean Eng. 234, 108795. doi: 10.1016/j.oceaneng.2021.108795
Meng F., Xu D., Song T. (2022). Atdnns: An adaptive time-frequency decomposition neural network-based system for tropical cyclone wave height real-time forecasting. Future Generation Comput. Syst 133, 297–306. doi: 10.1016/j.future.2022.03.029
Phillips O. M. (1957). On the generation of waves by turbulent wind. J. fluid mechanics 2, 417–445. doi: 10.1017/s0022112057000233
Shao W., Sheng Y., Li H., Shi J., Ji Q., Tan W., et al. (2018). Analysis of wave distribution simulated by wavewatch-iii model in typhoons passing beibu gulf, china. Atmosphere 9, 265. doi: 10.3390/atmos9070265
Shi X., Chen Z., Wang H., Yeung D.-Y., Wong W.-K., Woo W.-c. (2015). Convolutional lstm network: A machine learning approach for precipitation nowcasting. Adv. Neural Inf. Process. Syst 28. doi: 10.48550/arXiv.1506.04214
Song T., Jiang J., Li W., Xu D. (2020). A deep learning method with merged lstm neural networks for ssha prediction. IEEE J. Selected Topics Appl. Earth Observations Remote Sens. 13, 2853–2860. doi: 10.1109/jstars.2020.2998461
Song T., Li Y., Meng F., Xie P., Xu D. (2022). A novel deep learning model by bigru with attention mechanism for tropical cyclone track prediction in the northwest pacific. J. Appl. Meteorology Climatology 61, 3–12. doi: 10.1175/jamc-d-20-0291.1
Sverdrup H. U., Munk W. H. (1947). Wind, sea and swell: Theory of relations for forecasting Vol. 601 (Washington, D.C, United States: Hydrographic Office). doi: 10.5962/bhl.title.38751
Tolman H. L., et al. (2009). User manual and system documentation of wavewatch iii tm version 3.14. Tech. note MMAB Contribution 276. Available at: https://polar.ncep.noaa.gov/mmab/papers/tn276/MMAB_276.pdf
Van Aartrijk M. L., Tagliola C. P., Adriaans P. W. (2002). Ai on the ocean: The robosail project. In ECAI (Citeseer) 133, 653–657. Available at: www.robosail.com/research/the_RoboSail_project.pdf
Wei J., Malanotte-Rizzoli P., Eltahir E. A. B., Xue P., Xu D. (2013). Coupling of a regional atmospheric model (regcm3) and a regional oceanic model (fvcom) over the maritime continent. Climate Dynamics 43, 1575–1594. doi: 10.1007/s00382-013-1986-3
Zheng K., Sun J., Guan C., Shao W. (2016). Analysis of the global swell and wind sea energy distribution using wavewatch iii. Adv. Meteorology . 2016 doi: 10.1155/2016/8419580
Zhou S., Xie W., Lu Y., Wang Y., Zhou Y., Hui N., et al. (2021). Convlstm-based wave forecasts in the south and east china seas. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.680079
Keywords: wave height forecast, deep learning, high spatial and temporal resolution, new mechanism, long time prediction
Citation: Song T, Han R, Meng F, Wang J, Wei W and Peng S (2022) A significant wave height prediction method based on deep learning combining the correlation between wind and wind waves. Front. Mar. Sci. 9:983007. doi: 10.3389/fmars.2022.983007
Received: 30 June 2022; Accepted: 12 September 2022; Published: 03 October 2022.
Reviewed by:
Copyright © 2022 Song, Han, Meng, Wang, Wei and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiqiu Peng, [email protected] ; Fan Meng, [email protected]
This article is part of the Research Topic
Spatiotemporal Modeling and Analysis in Marine Science
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- J Healthc Eng
- v.2021; 2021

Machine Learning Based Diabetes Classification and Prediction for Healthcare Applications
Umair muneer butt.
1 School of Computer Sciences, Universiti Sains Malaysia, Penang, Malaysia
Sukumar Letchmunan
Mubashir ali.
2 Department of Management,Information and Production Engineering, University of Bergamo, Bergamo, Italy
Fadratul Hafinaz Hassan
Anees baqir.
3 Department of Environmental Sciences,Informatics,and Statistics, Ca'Foscari University of Venice, Venice, Italy
Hafiz Husnain Raza Sherazi
4 School of Computing and Engineering, University of West London, London, UK
Associated Data
The data used to support the findings of this study are included within the article.
The remarkable advancements in biotechnology and public healthcare infrastructures have led to a momentous production of critical and sensitive healthcare data. By applying intelligent data analysis techniques, many interesting patterns are identified for the early and onset detection and prevention of several fatal diseases. Diabetes mellitus is an extremely life-threatening disease because it contributes to other lethal diseases, i.e., heart, kidney, and nerve damage. In this paper, a machine learning based approach has been proposed for the classification, early-stage identification, and prediction of diabetes. Furthermore, it also presents an IoT-based hypothetical diabetes monitoring system for a healthy and affected person to monitor his blood glucose (BG) level. For diabetes classification, three different classifiers have been employed, i.e., random forest (RF), multilayer perceptron (MLP), and logistic regression (LR). For predictive analysis, we have employed long short-term memory (LSTM), moving averages (MA), and linear regression (LR). For experimental evaluation, a benchmark PIMA Indian Diabetes dataset is used. During the analysis, it is observed that MLP outperforms other classifiers with 86.08% of accuracy and LSTM improves the significant prediction with 87.26% accuracy of diabetes. Moreover, a comparative analysis of the proposed approach is also performed with existing state-of-the-art techniques, demonstrating the adaptability of the proposed approach in many public healthcare applications.
1. Introduction
Public health is a fundamental concern for protecting and preventing the community from health hazard diseases [ 1 ]. Governments are spending a considerable amount of their gross domestic product (GDP) for the welfare of the public, and initiatives such as vaccination have prolonged the life expectancy of people [ 2 ]. However, for the last many years, there has been a considerable emergence of chronic and genetic diseases affecting public health. Diabetes mellitus is one of the extremely life-threatening diseases because it contributes to other lethal diseases, i.e., heart, kidney, and nerve damage [ 3 ].
Diabetes is a metabolic disorder that impairs an individual's body to process blood glucose, known as blood sugar. This disease is characterized by hyperglycemia resulting from defects in insulin secretion, insulin action, or both [ 3 ]. An absolute deficiency of insulin secretion causes type 1 diabetes (T1D). Diabetes drastically spreads due to the patient's inability to use the produced insulin. It is called type 2 diabetes (T2D) [ 4 ]. Both types are increasing rapidly, but the ratio of increase in T2D is higher than T1D. 90 to 95% of cases of diabetes are of T2D.
Inadequate supervision of diabetes causes stroke, hypertension, and cardiovascular diseases [ 5 ]. To avoid and reduce the complications due to diabetes, a monitoring method of BG level plays a prominent role [ 6 ]. A combination of biosensors and advanced information and communication technology (ICT) provides an efficient real-time monitoring management system for the health condition of diabetic patients by using SMBG (self-monitoring of blood glucose) portable device. A patient can check the changes in glucose level in his blood by himself [ 7 ]. Users can better understand BG changes by using CGM (continuous glucose monitoring) sensors [ 4 ].
By exploiting the advantages of the advancement in modern sensor technology, IoT, and machine learning techniques, we have proposed an approach for the classification, early-stage identification, and prediction of diabetes in this paper. The primary objective of this study is twofold. First, to classify diabetes into predefined categories, we have employed three widely used classifiers, i.e., random forest, multilayer perceptron, and logistic regression. Second, for the predictive analysis of diabetes, long short-term memory (LSTM), moving averages (MA), and linear regression (LR) are used. To demonstrate the effectiveness of the proposed approach, PIMA Indian Diabetes is used for experimental evaluation. We concluded that, in experimental evaluation, MLP achieved an accuracy of 86.083% in diabetes classification as compared to the other classifiers and LSTM achieved a prediction accuracy of 87.26% for the prediction of diabetes. Moreover, we have also performed a comparative analysis of the proposed approach with existing state-of-the-art approaches. The accuracy results of our proposed approach demonstrate its adaptability in many healthcare applications.
Besides, we have also presented the IoT-based hypothetical diabetes self-monitoring system that uses BLE (Bluetooth Low Energy) devices and data processing in real-time. The latter technique used two applications: Apache Kafka (for streaming messages and data) and MongoDB (to store data). By utilizing BLE-based sensors, one can collect essential sign data about weight and blood glucose. These data will be handled by data processing techniques in a real-time environment. A BLE device will receive all the data produced by sensors and other necessary information about the patient that resides in the user application, installed on the cell phone. The raw data produced by sensors will be processed using the proposed approach to produce results, suggestions, and treatment from the patient's server-side.
The rest of the paper is organized as follows. In Section 2 , the paper presents the motivations for the proposed system by reviewing state-of-the-art techniques and their shortcomings. It covers the literature review about classification, prediction, and IoT-based techniques for healthcare. Section 3 highlights the role of physical activity in diabetes prevention and control. In Section 4 , we proposed the design and architecture of the diabetes classification and prediction systems. Section 5 discusses the results and performance of the proposed approach with state-of-the-art techniques. In Section 6 , an IoT-based hypothetical system is presented for real-time monitoring of diabetes. Finally, the paper is concluded in Section 7 , outlining the future research directions.
2. Literature Review
In this section, we discussed the classification and prediction algorithms for diabetes prediction in healthcare. Particularly, the significance of BLE-based sensors and machine learning algorithms is highlighted for self-monitoring of diabetes mellitus in healthcare. Machine learning plays an essential part in the healthcare industry by providing ease to healthcare professionals to analyze and diagnose medical data [ 8 – 12 ]. Moreover, intelligent healthcare systems are providing real-time clinical care to needy patients [ 13 , 14 ]. The features covered in this study are compared with the state-of-the-art studies ( Table 1 ).
Features' comparison of the proposed study vs. state-of-the-art studies.
2.1. Diabetes Classification for Healthcare
Health condition diagnosis is an essential and critical aspect for healthcare professionals. Classification of a diabetes type is one of the most complex phenomena for healthcare professionals and comprises several tests. However, analyzing multiple factors at the time of diagnosis can sometimes lead to inaccurate results. Therefore, interpretation and classification of diabetes are a very challenging task. Recent technological advances, especially machine learning techniques, are incredibly beneficial for the healthcare industry. Numerous techniques have been presented in the literature for diabetes classification.
Qawqzeh et al. [ 15 ] proposed a logistic regression model based on photoplethysmogram analysis for diabetes classification. They used 459 patients' data for training and 128 data points to test and validate the model. Their proposed system correctly classified 552 persons as nondiabetic and achieved an accuracy of 92%. However, the proposed technique is not compared with state-of-the-art techniques. Pethunachiyar [ 16 ] presented a diabetes mellitus classification system using a machine learning algorithm. Mainly, he used a support vector machine with different kernel functions and diabetes data from the UCI Machine Repository. He found SVM with linear function more efficient than naïve Bayes, decision tree, and neural networks. Nevertheless, the state-of-the-art comparison is missing and parameter selection is not elaborated.
Gupta et al. [ 17 ] exploited naïve Bayes and support vector machine algorithms for diabetes classification. They used the PIMA Indian Diabetes dataset. Besides, they used a feature selection based approach and k-fold cross-validation to improve the accuracy of the model. The experimental results showed the supremacy of the support vector machine over the naïve Bayes model. However, state-of-the-art comparison is missing along with achieved accuracy. Choubey et al. [ 18 ] presented a comparative analysis of classification techniques for diabetes classification. They used PIMA Indian data collected from the UCI Machine Learning Repository and a local diabetes dataset. They used AdaBoost, K-nearest neighbor regression, and radial basis function to classify patients as diabetic or not from both datasets. Besides, they used PCA and LDA for feature engineering, and it is concluded that both are useful with classification algorithms for improving accuracy and removing unwanted features.
Maniruzzaman et al. [ 19 ] used a machine learning paradigm to classify and predict diabetes. They utilized four machine learning algorithms, i.e., naive Bayes, decision tree, AdaBoost, and random forest, for diabetes classification. Also, they used three different partition protocols along with the 20 trials for better results. They used US-based National Health and Nutrition Survey data of diabetic and nondiabetic individuals and achieved promising results with the proposed technique. Ahuja et al. [ 20 ] performed a comparative analysis of various machine learning approaches, i.e., NB, DT, and MLP, on the PIMA dataset for diabetic classification. They found MLP superior as compared to other classifiers. The authors suggested that the performance of MLP can be enhanced by fine-tuning and efficient feature engineering. Recently, Mohapatra et al. [ 21 ] have also used MLP to classify diabetes and achieved an accuracy of 77.5% on the PIMA dataset but failed to perform state-of-the-art comparisons. MLP has been used in the literature for various healthcare disease classifications such as cardiovascular and cancer classification [ 35 , 36 ].
2.2. Predictive Analysis of Diabetes for Healthcare
Accurate classification of diabetes is a fundamental step towards diabetes prevention and control in healthcare. However, early and onset identification of diabetes is much more beneficial in controlling diabetes. The diabetes identification process seems tedious at an early stage because a patient has to visit a physician regularly. The advancement in machine learning approaches has solved this critical and essential problem in healthcare by predicting disease. Several techniques have been proposed in the literature for diabetes prediction.
Singh and Singh [ 22 ] proposed a stacking-based ensemble method for predicting type 2 diabetes mellitus. They used a publicly available PIMA dataset from the UCI Machine Learning Repository. The stacking ensemble used four base learners, i.e., SVM, decision tree, RBF SVM, and poly SVM, and trained them with the bootstrap method through cross-validation. However, variable selection is not explicitly mentioned and state-of-the-art comparison is missing.
Kumari et al. [ 23 ] presented a soft computing-based diabetes prediction system that uses three widely used supervised machine learning algorithms in an ensemble manner. They used PIMA and breast cancer datasets for evaluation purposes. They used random forest, logistic regression, and naïve Bayes and compared their performance with state-of-the-art individual and ensemble approaches, and their system outperforms with 79% accuracy.
Islam et al. [ 24 ] utilized data mining techniques, i.e., random forest, logistic regression, and naïve Bayes algorithm, to predict diabetes at the early or onset stage. They used 10-fold cross-validation and percentage split techniques for training purposes. They collected diabetic and nondiabetic data from 529 individuals directly from a hospital in Bangladesh through questionnaires. The experimental results show that random forest outperforms as compared to other algorithms. However, the state-of-the-art comparison is missing and achieved accuracy is not reported explicitly.
Malik et al. [ 25 ] performed a comparative analysis of data mining and machine learning techniques in early and onset diabetes mellitus prediction in women. They exploited traditional machine learning algorithms for proposing a diabetes prediction framework. The proposed system is evaluated on a diabetes dataset of a hospital in Germany. The empirical results show the superiority of K-nearest neighbor, random forest, and decision tree compared to other traditional algorithms.
Hussain and Naaz [ 26 ] presented a thorough review of machine learning models presented during 2010–2019 for diabetes prediction. They compared traditional supervised machine learning models with neural network-based algorithms in terms of accuracy and efficiency. They used Matthews correlation coefficient for evaluation purposes and observed naïve Bayes and random forest's supremacy compared to other algorithms.
2.3. Real-Time IoT-Based Processing of Healthcare Data
Real-time diabetes prediction is a complicated task. The emerging use of sensors in healthcare paved the path to handle fatal diseases [ 37 ]. Several techniques have been presented in the literature to classify and predict diabetes. Acciaroli et al. [ 4 ] exposed two accurate meters to measure diabetes in blood with less error rate. Furthermore, these commercial versions of glucometers are Accu-Chek with 6.5% error and CareSens with 4.0% error. Buckingham et al. [ 38 ] described the accuracy link of CGM with the calibration sensor. Alfian et al. [ 27 ] uncovered that the FDA had accepted CGM sensors for monitoring glucose in different trends and patterns. Moreover, at one particular time, one glucose reading should not be used to analyze the amount of insulin as not accepted in a glucometer. Rodríguez et al. [ 28 ] proposed a structural design containing a local gateway as a smartphone, cloud system, and sensors for advanced management of diabetes.
Filippoupolitis et al. [ 29 ] planned action to acknowledge a system using Bluetooth Low Energy (BLE) beacons and smartwatches. Mokhtari et al. considered technologies working with BLE for activity labeling and resident localization [ 30 ]. Gentili et al. [ 31 ] have used BLE with another application called Blue Voice, which can reveal the probability of multimedia communication of sensor devices and speech streaming service. Suárez et al. [ 32 ] projected a monitoring system based on the BLE device for air quality exposure with the environmental application. It aims at defining potential policy responses and studies the variables that are interrelated between societal level factors and diabetes prevalence [ 33 , 34 ].
Wang et al. [ 39 ] have given a general idea of the up-to-date BLE technology for healthcare systems based on a wearable sensor. They suggested that low-powered communication sensor technologies such as a BLE device can make it feasible for wearable systems of healthcare because it can be used without location constraints and is light in weight. Moreover, BLE is the first wireless technology in communication for healthcare devices in the form of a wearable device that meets expected operating requirements with low power, communication with cellular directly, secure data transmission, interoperability, electronic compatibility, and Internet communications. Rachim and Chung [ 40 ] have suggested one transmission system that used deficient power to observe the heart's activity through electrocardiograph signals using a BLE device for data transmission collecting by armband sensors and smartphones.
Mora et al. projected a dispersed structure using the IoT model to check human biomedically generated signals in reports using a BLE sensor device [ 41 ]. Cappon et al. [ 42 ] explored the study of CGM wearable sensors' prototypes and features of the commercial version currently used. Årsand et al. [ 43 ] offered the easiest method for monitoring blood glucose, physical activity, insulin injections, and nutritional information using smartphones and smartwatches. Morón et al. [ 44 ] observed the performance of the smartphone used in the medical field. Lee and Yoo [ 45 ] anticipated a structure using PDA (personal digital assistant) to manage diabetic patient's conditions better. It can also be used to send information about blood pressure, BG level, food consumption, and exercise plan of a patient with diabetes and give the direction of treatment by monitoring physical activity, food consumption, and insulin prescribed amount.
Rodríguez et al. [ 28 ] suggested an application for the smartphone, which can be used to receive the data from the sensor using a glucometer automatically. Rodríguez-Rodríguez et al. [ 46 ] suggested that checking the patient's glucose level and heart rate using sensors will produce colossal data, and analysis on big data can be used to solve this problem.
3. Role of Physical Activity in Prevention and Control of Diabetes Mellitus
Generally, physical activity is the first prevention and control strategy suggested by healthcare professionals to diabetic or prediabetic patients [ 47 ]. Among diet and medicine, exercise is a fundamental component in diabetes, cardiovascular disease, obesity, and lifestyle rescue programs. Nonetheless, dealing with all the fatal diseases has a significant economic burden. However, diabetes mellitus emerged as a devastating problem for the health sector and economy of a country of this century.
Recently, the international diabetes prevention and control federation predicts that diabetes can affect more than 366 million people worldwide [ 49 ]. The disease control and prevention center in the US alarmed the government that diabetes can affect more than 29 million people [ 50 ]. While these alarming numbers are continuously increasing, they will burden the economy around the globe. Therefore, researchers and healthcare professionals worldwide are researching and proposing guidelines to prevent and control this life-threatening disease. Sato [ 51 ] presented a thorough survey on the importance of exercise prescription for diabetes patients in Japan. He suggested that prolonged sitting should be avoided and physical activity should be performed every 30 minutes. Kirwan et al. [ 47 ] emphasized regular exercise to control and prevent type 2 diabetes. Particularly, they studied the metabolic effect on tissues of diabetic patients and found very significant improvements in individuals performing regular exercise. Moser et al. [ 48 ] have also highlighted the significance of regular exercise in improving the functionality of various organs of the body, as shown in Figure 1 .

Impact of regular exercise on metabolism of diabetic patients [ 48 ].
Yang et al. [ 52 ] focused on exercise therapy which plays a significant role in treating diabetes and its associated side effects. Specifically, they discovered cytokines which gives a novel insight into diabetes control, but the sequence is still under study. Kim and Jeon [ 53 ] presented a systematic overview of the effect of different exercises on the metabolism improvement of diabetic young individuals. They pointed out that several studies reported the significance of exercise on insulin, BP, and BG level improvement. However, none of these studies mentions the beta-cell improvement. Therefore, many challenges persist in diabetes prevention and control, which need serious attention from researchers worldwide.
4. Proposed Diabetic Classification and Prediction System for Healthcare
The proposed diabetes classification and prediction system has exploited different machine learning algorithms. First, to classify diabetes, we utilized logistic regression, random forest, and MLP. Notably, we fine-tuned MLP for classification due to its promising performance in healthcare, specifically in diabetes prediction [ 20 , 21 , 35 , 36 ]. The proposed MLP architecture and algorithm are shown in Figure 2 and Algorithm 1 , respectively.

Proposed MLP architecture with eight variables as input for diabetes classification.

Diabetes classification algorithm using MLP for healthcare.
Second, we implement three widely used machine learning algorithms for diabetes prediction, i.e., moving averages, linear regression, and LSTM. Mainly, we optimized LSTM for crime prediction due to its outstanding performance in real-world applications, particularly in healthcare [ 53 ]. The implementation details of the proposed algorithms are as follows.
4.1. Diabetes Classification Techniques
For diabetic classification, we fine-tuned three widely used state-of-the-art techniques. Mainly, a comparative analysis is performed among the proposed techniques for classifying an individual in either of the diabetes categories. The details of the proposed diabetes techniques are as follows.
4.1.1. Logistic Regression
It is appropriate to use logistic regression when the dependent variable is binary [ 54 ], as we have to classify an individual in either type 1 or type 2 diabetes. Besides, it is used for predictive analysis and explains the relationship between a dependent variable and one or many independent variables, as shown in equation ( 1 ). Therefore, we used the sigmoid cost function as a hypothesis function ( h θ ( x )). The aim is to minimize cost function J ( θ ). It always results in classifying an example either in class 1 or class 2.
4.1.2. Random Forest (RF)
As its name implies, it is a collection of models that operate as an ensemble. The critical idea behind RF is the wisdom of the crowd, each model predicts a result, and in the end, the majority wins. It has been used in the literature for diabetic prediction and was found to be effective [ 55 ]. Given a set of training examples X = x 1 , x 2 ,…, x m and their respective targets Y = y 1 , y 2 ,…, y m , RF classifier iterates B times by choosing samples with replacement by fitting a tree to the training examples. The training algorithm consists of the following steps depicted in equation ( 2 ).
- For b = 1... B , sample with replacement n training examples from X and Y .
- Train a classification tree f b on X b and Y b .
4.1.3. Multilayer Perceptron
For diabetes classification, we have fine-tuned multilayer perceptron in our experimental setup. It is a network where multiple layers are joined together to make a classification method, as shown in Figure 2 . The building block of this model is perceptron, which is a linear combination of input and weights. We used a sigmoid unit as an activation function shown in Algorithm 1 . The proposed algorithm consists of three main steps. First, weights are initialized and output is computed at the output layer ( δ k ) using the sigmoid activation function. Second, the error is computed at hidden layers ( δ h ) for all hidden units. Finally, in a backward manner, all network weights ( w i , j ) are updated to reduce the network error. The detailed procedure is outlined in Algorithm 1 for diabetes classification.
Figure 2 shows the multilayer perceptron classification model architecture where eight neurons are used in the input layer because we have eight different variables. The middle layer is the hidden layer where weights and input will be computed using a sigmoid unit. In the end, results will be computed at the output layer. Backpropagation is used for updating weights so that errors can be minimized for predicting class labels. For simplicity, only one hidden layer is shown in the architecture, which in reality is much denser.
Input data from the input layer are computed on the hidden layers with the input values and weights initialized. Every unit in the middle layer called the hidden layer takes the net input, applies activation function “sigmoid” on it, and transforms the massive data into a smaller range between 0 and 1. The calculation is functional for every middle layer. The same procedure is applied on the output layer, which leads to the results towards the prediction for diabetes.
4.2. Diabetes Prediction
It is more beneficial to identify the early symptoms of diabetes than to cure it after being diagnosed. Therefore, in this study, a diabetes prediction system is proposed where three state-of-the-art machine learning algorithms are exploited, and a comparative analysis is performed. The details of the proposed approaches are as follows.
4.2.1. Moving Averages
To predict diabetes, we used moving averages with the experimental setup due to its effectiveness in diabetes prediction for children [ 56 ]. It is based on a calculation that analyzes data points by creating a series of averages of the subset of the data randomly. The moving average algorithm is based on the “forward shifting” mechanism. It excludes the first number from the series and includes the next value in the dataset, as shown in equation ( 3 ). The input values are calculated by averaging ( P SM ) the train data at certain time stamps P M + P M + … P M −( n −1) . The algorithm used past observations as input and predicted future events.
4.2.2. Linear Regression
Second, a linear regression model is applied to the PIMA Indian dataset with the same experimental setup. We used this approach to model a relationship between a dependent variable, that is, outcome in our case, and one or more independent variables. The autonomous variable response affects a lot on the target/dependent variable, as shown in equation ( 4 ). We use a simplified hypothesis and cost function for multivariate linear regression, as there are eight different variables in our dataset [ 57 ]. We choose a very simplified hypothesis function ( h θ ( x )). The aim is to minimize cost function J ( θ ) by choosing the suitable weight ( θ T x ) parameters and minimizing sum of squared error (SSE).
4.2.3. Long Short-Term Memory
For diabetic forecasting, we have calibrated the long short-term memory algorithm with our experimental setup. The proposed approach outperformed as compared to other state-of-the-art techniques implemented, as shown in Table 2 . LSTM is based on recurrent neural network (RNN) architecture, and it has feedback connections that make it suitable for diabetes forecasting [ 58 ]. LSTM mainly consists of a cell, keep gate, write gate, and an output gate, as shown in Figure 3 . The key behind using LSTM for this problem is that the cell remembers the patterns over a long period, and three portals help regulate the information flow in and out of the system. The details are presented in Algorithm 2 .

BG prediction using long short-term memory (LSTM) algorithm.

Diabetes prediction algorithm by exploiting LSTM for healthcare.
Performance comparison of classifiers in diabetes classification.
Input to the algorithm is eight attributes enlisted in Table 3 , measured from healthy and diabetic patients. The proposed LSTM-based diabetes prediction algorithm is trained with 80% of the data, and the remaining 20% is used for testing. We fine-tuned the prediction model by using a different number of LSTM units in the cell state. This fine-tuning helps to identify more prominent features in the dataset. These features will be kept in the cell state of the keep gate of the LSTM and will be given more weightage because they provide more insights to predict BG level. After that, we updated the network's weights by pointwise addition of the cell state and passed only those essential attributes for BG prediction. At this stage, we captured the dependencies between diabetes parameters and the output variable. Finally, the output gate updates the cell state and outputs/forwards only those variables that can be mapped efficiently on the outcome variable.
Description of variables in the dataset.
The diabetes prediction algorithm consists of three fundamental steps. First, weights are initialized and a sigmoid unit is used in the forget/keep gate to decide which information should be retained from previous and current inputs ( C t −1 , h t −1 , and x t ). The input/write gate takes the necessary information from the keep gate and uses a sigmoid unit which outputs a value between 0 and 1. Besides, a Tan h unit is used to update the cell state C t and combine both outputs to update the old cell state to the new cell state.
Finally, inputs are processed at the output gate and again a sigmoid unit is applied to decide which cell state should be output. Also, Tan h is applied to the incoming cell state to push the output between 1 and −1. If the output of the gate is 1, then the memory cell is still relevant to the required production and should be kept for future results. If the output of the gate is 0, the memory cell is not appropriate, so it should be erased. For the write gate, the suitable pattern and type of information will be determined written into the memory cell. The proposed LSTM model predicts the BG level ( h t ) as output based on the patient's existing BG level ( X t ).
5. Experimental Studies
The proposed diabetes classification and prediction algorithm is evaluated on a publicly available PIMA Indian Diabetes dataset ( https://www.niddk.nih.gov/health-information/diabetes ). Besides, a comparative analysis is performed with state-of-the-art algorithms. The experimental results show the supremacy of the proposed algorithm as compared to state-of-the-art algorithms. The details of the dataset, performance measures, and comparative analysis performed are described in the following sections.
5.1. Dataset
This study used the PIMA Indian Diabetes (PID) dataset taken from the National Institute of Diabetes and Kidney Diseases center [ 59 ]. The primary objective of using this dataset is to build an intelligent model that can predict whether a person has diabetes or not, using some measurements included in the dataset. There are eight medical predictor variables and one target variable in the dataset. Diabetes classification and prediction are a binary classification problem. The details of the variables are shown in Table 3 .
The dataset consists of 768 records of different healthy and diabetic female patients of age greater than twenty-one, as shown in Figure 4 . The feature value distribution is shown in Figure 5 . The target variable outcome contains only two values, 0 and 1. The primary objective of using this dataset was to predict diabetes diagnostically. Whether a user has a chance of diabetes in the coming four years in women belongs to PIMA Indian. The dataset has a total of eight variables: glucose tolerance, no. of pregnancies, body mass index, blood pressure, age, insulin, and Diabetes Pedigree Function. All eight attributes shown in Table 3 are used for the training dataset in the classification model in this work.

PIMA data distribution.

Dataset features' distribution visualization.
5.2. Experimental Result and Discussion
This paper compares the proposed diabetes classification and prediction system with state-of-the-art techniques using the same experimental setup on the PIMA Indian dataset. The following sections highlighted the performance measure used and results attained for classification and prediction, and a comparative analysis with baseline studies is presented.
5.2.1. Performance Metrics
Three widely used state-of-the-art performance measures (Recall, Precision, and Accuracy) are used to evaluate the performance of proposed techniques, as shown in Table 4 . TP shows a person does not have diabetes and identified as a nondiabetic patient, and TN shows a diabetic patient correctly identified as a diabetic patient. FN shows the patient has diabetes but is predicted as a healthy person. Moreover, FP shows the patient is a healthy person but predicted as a diabetic patient. The algorithm utilized 10-fold cross-validation for training and testing the classification and prediction model.
Performance metrics for diabetes classification.
For diabetes prediction, the two most commonly used performance measures are the means correlation coefficient ( r /Pearson R ) and root mean square error (RMSE), as shown in Table 5 . R is mainly used to measure the linear dependence strength among the two variables. One variable is for actual value, and another variable is for predicted values. RMSE generates a hint of the overall correctness of the estimate. There can be three values for correlation: 0 for no relation, 1 for positive correlation, and −1 for the negative correlation. RMSE shows the difference between actual values and predicted values.
Performance measure for diabetes prediction.
5.2.2. Attained Results of Diabetic Classification Technique
For diabetic classification, three state-of-the-art classifiers are evaluated on the PIMA dataset. The results illustrate that the fine-tuned MLP algorithm obtained the highest accuracy of 86.083% as compared to state-of-the-art systems, as shown in Table 2 .
It is evident from the results that our proposed calibrated MLP model could be used for the effective classification of diabetes. The proposed classification approach can also be beneficial in the future with our proposed hypothetical system. Data of weight scales, blood pressure monitor, and blood glucometer will be collected through sensor devices such as BLE and input of user's demographic data (for example, date of birth, height, and age). The proposed MLP algorithm outperforms with 86.6% Precision, 85.1% Recall, and 86.083% Accuracy, as shown in Figure 6 . These results are outstanding for decision-making with the proposed hypothetical system to determine patient diabetes, T1D or T2D.

Performance comparison of classifiers.
We also have explored the dataset used in Andy Choens' study [ 27 ]. This dataset consists of records of only one patient. The information was recorded every five minutes. The collection of data was made by using a sensor device (a CGM device). This device allows the patient to store information about BG every five minutes. So, the recorded data by using this device are in massive amounts. Dataset was limited, and most data were noisy that can affect the accuracy of the proposed system, so we neglected it.
5.2.3. Achieved Results of Diabetic Prediction Techniques
For diabetic prediction, we implemented three state-of-the-art algorithms, i.e., linear regression, moving averages, and LSTM. Notably, we fine-tuned LSTM and compared its performance with other algorithms. It is evident from Figure 7 and Table 6 that the LSTM outperformed as compared to other algorithms implemented in this study.

Performance comparison of forecasting model.
Forecasting model comparison for BG.
Table 2 shows the performance values of prediction models with RMSE and r evaluation measures. The proposed fine-tuned LSTM produced the highest accuracy, 87.26%, compared to linear regression and moving average. We can see in Table 6 that the correlation coefficient value is 0.999 using LSTM, −0.071 for linear regression, and 0.710 for moving average, as shown in Figure 7 .
5.2.4. Comparison of the Proposed Method with Baseline Studies
Different baseline studies have been implemented and compared with the proposed system to verify the performance of the proposed diabetes classification and prediction system. Mainly, we focus on those studies that used the PIMA dataset.
First, we compare the state-of-the-art diabetes classification techniques with the proposed technique. All the baseline techniques [ 17 – 19 ] used the PIMA dataset and the same evaluation measures used in this study. In particular, the authors compared naïve Bayes [ 17 ], PCA_CVR (classification via regression) [ 18 ], and SVM [ 19 ] with different machine learning techniques for diabetes classification. However, the proposed fine-tuned MLP-based diabetes classification technique outperformed as compared to baseline studies, as shown in Figure 8 .

Proposed diabetes classification method vs. state-of-the-art techniques.
Several attempts have also been made in the literature for diabetic prediction due to its importance in real life. For this comparison, we have chosen the most recent and state-of-the-art techniques. We compare the proposed system performance with the recent state-of-the-art systems [ 60 – 65 ], as shown in Figure 9 and Table 7 . The proposed method outperformed as compared to state-of-the-art systems with an accuracy of 87.26%, all the compared systems evaluated on the PID with the same experimental setup.

Proposed diabetes prediction method vs. state-of-the-art systems.
Proposed prediction method vs. state-of-the-art systems.
6. Proposed Hypothetical IoT-Based Diabetic Monitoring System for Healthcare
This study has also proposed the architecture of a hypothetical diabetic monitoring system for diabetic patients. The proposed hypothetical system will enable a patient to control, monitor, and manage their chronic conditions in a better way at their homes. The monitoring system will store the health activities and create interaction between patients, smartphones, sensor medical devices, web servers, and medical teams by providing a platform having wireless communication devices, as shown in Figure 10 . The central theme of the proposed healthcare monitoring system is the collection of data from sensors using wireless devices and transmitting to a remote server for diagnosis and treatment of diabetes. Knowledge-based data are stored. Rule-based procedures will be applied for the suggestions and treatment of diabetes, informing the patient about his current health condition, prediction, and recommendation of future changes in BG.

The proposed hypothetical architecture of the healthcare monitoring system.
First, essential data about patient health will be collected from sensors such as BLE wireless devices. Data comprised weight, blood pressure, blood glucose, and heartbeat, along with some demographic information such as age, sex, name, and CNIC (Social Security Number). Some information is required in the application installed on the user's mobile and sensor data. All completed data in the application will be transferred to the real-time data processing system. On the other side, aggregate data will be stored in MongoDB for future processing. Analysis and prepossessing techniques are performed to extract rules from the knowledge base for the treatment and suggestions about the user. Results and treatment procedures will be sent to the monitoring system, and finally, the user can get the output by interacting with their android mobile phone. In the end, the patient will know about the health condition and risk prediction of diabetes based on the data transferred by their application and stored data from history about the user.
6.1. Tools and Technology for Implementation of Hypothetical System for Healthcare
The proposed structural design for hypothetical real-time processing and monitoring of diabetes is shown in Figure 11 . The data from the user's mobile will be transmitted in the JavaScript Object Notation (JSON) format to the Application Program Interface (API) in any language. The data produced at this stage will be in the form of messages, which are then transferred to the Kafka application [ 27 ]. Kafka will store all the data and messages and deliver the required data and processed output to the endpoints that could be a web server, monitoring system, or a database for permanent storage. In Kafka, application data are stored in different brokers, which can cause latency issues. Therefore, within the system architecture, it is vital to consider processing the readings from the sensors closer to the place where data are acquired, e.g., on the smartphone. The latency problem could be solved by placing sensors close to the place, such as a smartphone where data are sent and received.

Implementation level details of the proposed hypothetical system.
This inclusion will make the overall network architecture compliant to the emerging Edge and Fog computing paradigms, whose importance in critical infrastructures such as hospitals is gaining momentum. It is essential to consider the Edge and Fog computation paradigm while sending and receiving data from smartphones to increase the performance of the hypothetical system. Edge computing utilizes sensors and mobile devices to process, compute, and store data locally rather than cloud computing. Besides, Fog computing places resources near data sources such as gateways to improve latency problems [ 9 ].
Apache Kafka will be used in real time as a delivery agent for messages in a platform that allows fault-tolerant, tall throughput, and low-latency publication. The vital signs' data collected by the patients are placed using the JSON format and then transmitted using wireless devices with the help of an android application having HTTP along with REST API for the confined remote server for the design [ 28 ]. Moreover, Node.js for web design will be used as a REST API to collect sensor data. Kafka application will receive it in the form of streams of records.
The sensor data that comes from the Kafka application is continuously generated and stored on the server. In the proposed system, the MongoDB NoSQL database will be used for data storage due to its efficiency in handling and processing real-world data [ 29 ]. The stored diabetes patient data can be input into our proposed diabetes classification and prediction techniques to get useful insights.
7. Conclusion
In this paper, we have discussed an approach to assist the healthcare domain. The primary objective of this study is twofold. First, we proposed an MLP-based algorithm for diabetes classification and deep learning based LSTM for diabetes prediction. Second, we proposed an IOT-based hypothetical real-time diabetic monitoring system. The proposed theoretical diabetic monitoring system will use a smartphone, BLE-based sensor device, and machine learning based methods in the real-time data processing environment to predict BG levels and diabetes. The primary objective of the proposed system is to help the users monitor their vital signs using BLE-based sensor devices with the help of their smartphones.
Moreover, the proposed model will help the users to find out the risk of diabetes at a very early stage and help them gaining future predictions of their BG increase levels. For diabetic classification and prediction, MLP and LSTM are fine-tuned. The proposed approaches are evaluated on the PIMA Indian Diabetes dataset. Both approaches are compared with state-of-the-art approaches and outperformed with an accuracy of 86.083% and 87.26%, respectively.
As future work, we plan to implement the android application for the proposed hypothetical diabetic monitoring system with the proposed classification and prediction approaches. Genetic algorithms can also be explored with the proposed prediction mechanism for better monitoring [ 24 , 64 , 66 – 71 ].
Acknowledgments
This work was funded by the School of Computer Sciences, Universiti Sains Malaysia, Penang, Malaysia.
Data Availability
Conflicts of interest.
The authors declare that there are no conflicts of interest regarding the publication of this article.
- Open access
- Published: 13 May 2024
Neighborhood based computational approaches for the prediction of lncRNA-disease associations
- Mariella Bonomo 1 &
- Simona E. Rombo 1 , 2
BMC Bioinformatics volume 25 , Article number: 187 ( 2024 ) Cite this article
106 Accesses
Metrics details
Long non-coding RNAs (lncRNAs) are a class of molecules involved in important biological processes. Extensive efforts have been provided to get deeper understanding of disease mechanisms at the lncRNA level, guiding towards the detection of biomarkers for disease diagnosis, treatment, prognosis and prevention. Unfortunately, due to costs and time complexity, the number of possible disease-related lncRNAs verified by traditional biological experiments is very limited. Computational approaches for the prediction of disease-lncRNA associations allow to identify the most promising candidates to be verified in laboratory, reducing costs and time consuming.
We propose novel approaches for the prediction of lncRNA-disease associations, all sharing the idea of exploring associations among lncRNAs, other intermediate molecules (e.g., miRNAs) and diseases, suitably represented by tripartite graphs. Indeed, while only a few lncRNA-disease associations are still known, plenty of interactions between lncRNAs and other molecules, as well as associations of the latters with diseases, are available. A first approach presented here, NGH, relies on neighborhood analysis performed on a tripartite graph, built upon lncRNAs, miRNAs and diseases. A second approach (CF) relies on collaborative filtering; a third approach (NGH-CF) is obtained boosting NGH by collaborative filtering. The proposed approaches have been validated on both synthetic and real data, and compared against other methods from the literature. It results that neighborhood analysis allows to outperform competitors, and when it is combined with collaborative filtering the prediction accuracy further improves, scoring a value of AUC equal to 0966.
Availability
Source code and sample datasets are available at: https://github.com/marybonomo/LDAsPredictionApproaches.git
Peer Review reports
Introduction
More than \(98\%\) of the human genome consists of non-coding regions, considered in the past as “junk” DNA. However, in the last decades evidence has been shown that non-coding genome elements often play an important role in regulating various critical biological processes [ 1 ]. An important class of non-coding molecules which have started to receive great attention in the last few years is represented by long non-coding RNAs (lncRNAs), that is, RNAs not translated into functional proteins, and longer than 200 nucleotides.
LncRNAs have been found to interplay with other molecules in order to perform important biological tasks, such as modulating chromatin function, regulating the assembly and function of membraneless nuclear bodies, interfering with signalling pathways [ 2 , 3 ]. Many of these functions ultimately affect gene expression in diverse biological and physiopathological contexts, such as in neuronal disorders, immune responses and cancer. Therefore, the alteration and dysregulation of lncRNAs have been associated with the occurrence and progress of many complex diseases [ 4 ].
The discovery of novel lncRNA-disease associations (LDAs) may provide valuable input to the understanding of disease mechanisms at lncRNA level, as well as to the detection of disease biomarkers for disease diagnosis, treatment, prognosis and prevention. Unfortunately, verifying that a specific lncRNA may have a role in the occurrence/progress of a given disease is an expensive process, therefore the number of disease-related lncRNAs verified by traditional biological experiments is yet very limited. Computational approaches for the prediction of potential LDAs can effectively decrease the time and cost of biological experiments, allowing for the identification of the most promising lncRNA-disease pairs to be further verified in laboratory (see [ 5 ] for a comprehensive review on the topic). Such approaches often train predictive models on the basis of the known and experimentally validated lncRNA-disease pairs (e.g., [ 6 , 7 , 8 , 9 ]). In other cases, they rely on the analysis of lncRNAs related information stored in public databases, such as their interaction with other types of molecules (e.g., [ 10 , 11 , 12 , 13 , 14 , 15 ]). As an example, large amounts of lncRNA-miRNA interactions have been collected in public databases, and plenty of experimentally confirmed miRNA-disease associations are available as well. However, although non-coding RNA function and its association with human complex diseases have been widely studied in the literature (see [ 16 , 17 , 18 ]), how to provide biologists with more accurate and ready-to-use software tools for LDAs prediction is yet an open challenge, due to the specific characteristics of lncRNAs (e.g., they are much less characterized than other non-coding RNAs.)
We propose three novel computational approaches for the prediction of LDAs, relying on the use of known lncRNA-miRNA interactions (LMIs) and miRNA-disease associations (MDAs). In particular, we model the problem of LDAs prediction as a neighborhood analysis performed on tripartite graphs, where the three sets of vertices represent lncRNAs, miRNAs and diseases, respectively, and vertices are linked according to LMIs and MDAs. Based on the assumption that similar lncRNAs interact with similar diseases [ 12 ], the first approach proposed here (NGH) aims at identifying novel LDAs by analyzing the behaviour of lncRNAs which are neighbors , in terms of their intermediate relationships with miRNAs. The main idea here is that neighborhood analysis automatically guides towards the detection of similar behaviours, and without the need of using a-priory known LDAs for training. Therefore, differently than other approaches from the literature, those proposed here do not involve verified LDAs in the prediction step, thus avoiding possible biases due to the fact that the number and variety of verified LDAs is yet very limited. The second presented approach (CF) relies on collaborative filtering, applied on the basis of common miRNAs shared by different lncRNAs. We have also explored the combination of neighborhood analysis with collaborative filtering, showing that this notably improves the LDAs prediction accuracy. Indeed, the third approach we have designed (NGH-CF) boosts NGH with collaborative filtering, and it is the best performing one, although also NGH and CF have been able to reach high accuracy values across all the different considered validation tests. In particular, Fig. 1 summarizes the research flowchart explained above.
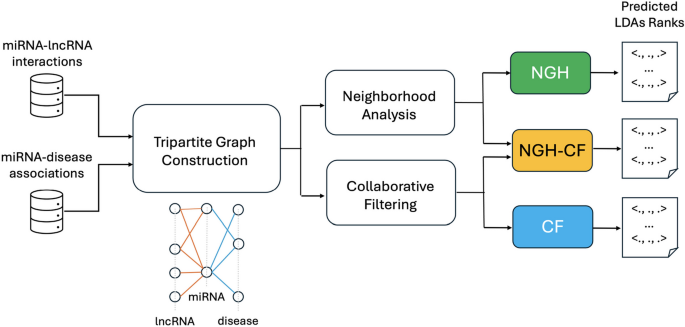
Flowchart of the research pipeline. The miRNA-lncRNA interactions and miRNA-disease associations are exploited for the construction of the tripartite graph. The tripartite graph, in its turn, is at the basis of both neighborhood analysis and collaborative filtering steps, from which the three proposed approaches are obtained: NGH from neighborhood analysis, CF from collaborative filtering, NGH-CF from the combination of the two ones. Each prediction approach returns in output a LDAs rank
The proposed approaches have been exhaustively validated on both synthetic and real datasets, and the result is that they outperform (also significantly) the other methods from the literature. The experimental analysis shows that the improvement in accuracy achieved by the methods proposed here is due to their ability in capturing specific situations neglected by competitors. Examples of that are represented by true LDAs, detected by our approaches and not by the other approaches in the literature, where the involved lncRNA does not present intermediate molecules in common with the associated disease, although its neighbor lncRNAs share a large number of miRNAs with that disease. Moreover, it is shown that our approaches are robust to noise obtained by perturbing a controlled percentage of lncRNA-miRNA interactions and miRNA-disease associations, with NGH-CF the best one also for robustness. The obtained experimental results show that the prediction methods proposed here may effectively support biologists in selecting significant associations to be further verified in laboratory.
Novel putative LDAs coming from the consensus of the three proposed methods, and not yet registered in the available databases as experimentally verified, are provided. Interestingly, the core of novel LDAs returned with highest score by all three approaches finds evidence in the recent literature, while many other high scored predicted LDAs involve less studied lncRNAs, thus providing useful insights for their better characterization.
A first group of approaches aim at using existing true validated cases to train the prediction system, in order to make it able to correctly detect novel cases.
In [ 19 ] a Laplacian Regularized Least Squares is proposed to infer candidates LDAs ( LRLSLDA ) by applying a semi-supervised learning framework. LRLSLDA assumes that similar diseases tend to correlate with functionally similar lncRNAs, and vice versa. Thus, known LDAs and lncRNA expression profiles are combined to prioritize disease-associated lncRNA candidates by LRLSLDA, which does not require negative samples (i.e., confirmed uncorrelated LDAs). In [ 20 ] the method SKF-LDA is proposed that constructs a lncRNA-disease correlation matrix, based on the known LDAs. Then, it calculates the similarity between lncRNAs and that between diseases, according to specific metrics, and integrates such data. Finally, a predicted LDA matrix is obtained by the Laplacian Regularized Least Squares method. The method ENCFLDA [ 6 ] combines matrix decomposition and collaborative filtering. It uses matrix factorization combined with elastic networks and a collaborative filtering algorithm, making the prediction model more stable and eliminating the problem of data over-fitting. HGNNLDA recently proposed in [ 21 ] is based on hypergraph neural network, where the associations are modeled as a lncRNA-drug bipartite graph to build lncRNA hypergraph and drug hypergraph. Hypergraph convolution is then used to learn correlation of higher-order neighbors from the lncRNA and drug hypergraphs. LDAI-ISPS proposed in [ 22 ] is a LDAs inference approach based on space projections of integrated networks, recostructing the disease (lncRNA) integrated similarities network via integrating multiple information, such as disease semantic similarities, lncRNA functional similarities, and known LDAs. A space projection score is finally obtained via vector projections of the weighted networks. In [ 7 ] a consensual prediction approach called HOPEXGB is presented, to identify disease-related miRNAs and lncRNAs by high-order proximity preserved embedding and extreme gradient boosting. The authors build a heterogeneous disease-miRNA-lncRNA (DML) information network by linking lncRNA, miRNA, and disease nodes based on their correlation, and generate a negative dataset based on the similarities between unknown and known associations, in order to reduce the false negative rate in the data set for model construction. The method MAGCNSE proposed in [ 23 ] builds multiple feature matrices based on semantic similarity and disease Gaussian interaction profile kernel similarity of both lncRNAs and diseases. MAGCNSE adaptively assigns weights to the different feature matrices built upon the lncRNAs and diseases similarities. Then, it uses a convolutional neural network to further extract features from multi-channel feature matrices, in order to obtain the final representations of lncRNAs and diseases that is used for the LDAs prediction task.
LDAFGAN [ 8 ] is a model designed for predicting associations between long non-coding RNAs (lncRNAs) and diseases. This method is based on a generative and a discriminative networks, typically implemented as multilayer fully connected neural networks, which generate synthetic data based on some underlying distribution. The generative and discriminative networks are trained together in an adversarial manner. The generative network tries to generate realistic representations of lncRNA-disease associations, while the discriminative network tries to distinguish between real and fake associations. This adversarial training process helps the generative network learn to generate more realistic associations. Once the model is trained, it can predict associations between new lncRNAs and diseases without requiring associated data for those specific lncRNAs. The model captures the data distribution during training, which enables it to make predictions even for unseen lncRNAs. The approach GCNFORMER [ 9 ] is based on graph convolutional network and transformer. First, it integrates the intraclass similarity and interclass connections between miRNAs, lncRNAs and diseases, building a graph adjacency matrix. Then, the method extracts the features between various nodes, by a graph convolutional network. To obtain the global dependencies between inputs and outputs, a transformer encoder with a multiheaded attention mechanism to forecast lncRNA-disease associations is finally applied.
As for the approaches summarized above, it is worth to point out that they may suffer of the fact that the experimentally verified LDAs are still very limited, therefore the training set may be rather incomplete and not enough diversified. For this reason, when such approaches are applied for de novo LDAs prediction, their performance may drastically go down [ 12 ].
Other approaches from the literature use intermediate molecules (e.g., miRNA) to infer novel LDAs. Such approaches are the most related to those we propose here.
The author in [ 11 ] proposes HGLDA , relying on HyperGeometric distribution for LDAs inference, that integrates MDAs and LMIs information. HGLDA has been successfully applied to predict Breast Cancer, Lung Cancer and Colorectal Cancer-related lncRNAs. NcPred [ 10 ] is a resource propagation technique, using a tripartite network where the edges associate each lncRNA with a disease through its targets. The algorithm proposed in [ 10 ] is based on a multilevel resource transfer technique, which computes the weights between each lncRNA-disease pair and, at each step, considers the resource transferred from the previous step. The approach in [ 24 ], referred to as LDA-TG for short in the following, is the antecedent of the approaches proposed here. It relies on the construction of a tripartite graph, built upon MDAs and LMIs. A score is assigned to each possible LDA ( l , d ) by considering both their respective interactions with common miRNAs, and the interactions with miRNAs shared by the considered disease d and other lncRNAs in the neighborhood of l on the tripartite graph. The approaches proposed here differ from LDA-TG for two main reasons. First, the score of LDA-TG is different from the one we introduce here, that allows to reach a better accuracy. Second, a further step based on collaborative filtering is considered here, which also improves the accuracy performance. A method for LDAs prediction relying on a matrix completion technique inspired by recommender systems is presented in [ 14 ]. A two-layer multi-weighted nearest-neighbor prediction model is adopted, using a method similar to memory-based collaborative filtering. Weights are assigned to neighbors for reassigning values to the target matrix, that is an adjacency matrix consisting of lncRNAs, diseases and miRNA. SSMF-BLNP [ 25 ] is based on the combination of selective similarity matrix fusion (SSMF) and bidirectional linear neighborhood label propagation (BLNP). In SSMF, self-similarity networks of lncRNAs and diseases are obtained by selective preprocessing and nonlinear iterative fusion. In BLNP, the initial LDAs are employed in both lncRNA and disease directions as label information for linear neighborhood label propagation.
A third category includes approaches based on integrative frameworks, proposed to take into account different types of information related to lncRNAs, such as their interactions with other molecules, their involvement in disorders and diseases, their similarities. This may improve the prediction step, taking into account simultaneously independent factors.
IntNetLncSim [ 26 ] relies on the construction of an integrated network that comprises lncRNA regulatory data, miRNA-mRNA and mRNA-mRNA interactions. The method computes a similarity score for all pairs of lncRNAs in the integrated network, then analyzes the information flow based on random walk with damping. This allows to infer novel LDAs by exploring the function of lncRNAs. SIMCLDA [ 12 ] identifies LDAs by using inductive matrix completion, based on the integration of known LDAs, disease-gene interactions and gene-gene interactions. The main idea in [ 12 ] is to extract feature vectors of lncRNAs and diseases by principal component analysis, and to calculate the interaction profile for a new lncRNA by the interaction profiles. MFLDA [ 27 ] is a Matrix Factorization based LDAs prediction model that first encodes directly (or indirectly) relevant data sources related to lncRNAs or diseases in individual relational data matrices, and presets weights for these matrices. Then, it simultaneously optimizes the weights and low-rank matrix tri-factorization of each relational data matrix. RWSF-BLP , proposed in [ 28 ], applies a random walk-based multi-similarity fusion method to integrate different similarity matrices, mainly based on semantic and expression data, and bidirectional label propagation. The framework LRWRHLDA is proposed in [ 15 ] based on the construction of a global multi-layer network for LDAs prediction. First, four isomorphic networks including a lncRNA similarity network, a disease similarity network, a gene similarity network and a miRNA similarity network are constructed. Then, six heterogeneous networks involving known lncRNA-disease, lncRNA-gene, lncRNA-miRNA, disease-gene, disease-miRNA, and gene-miRNA associations are built to design the multi-layer network. In [ 29 ] the LDAP-WMPS LDA prediction model is proposed, based on weight matrix and projection score. LDAP-WMPS consists on three steps: the first one computes the disease projection score; the second step calculates the lncRNA projection score; the third step fuses the disease projection score and the lncRNA projection score proportionally, then it normalizes them to get the prediction score matrix.
For most of the approaches summarized above, the performance is evaluated using the LOOCV framework, such that each known LDA is left out in turn as a test sample, and how well this test sample is ranked relative to the candidate samples (all the LDAs without the evidence to confirm their relationships) is computed.
The main goal of the research presented here is to provide more accurate computational methods for the prediction of novel LDAs, candidate for experimental validation in laboratory. To this aim, external information on both molecular interactions (e.g., lncRNA-miRNA interactions) and genotype-phenotype associations (e.g., miRNA-disease associations) is assumed to be available. Indeed, while only a restricted number of validated LDAs is yet available, large amounts of interactions between lncRNAs and other molecules (e.g., miRNAs, genes, proteins), as well as associations between these other molecules and diseases, are known and annotated in curated databases.
A commonly recognized assumption is that lncRNAs with similar behaviour in terms of their molecular interactions with other molecules, may also reflect such a similarity for their involvement in the occurrence and progress of disorders and diseases [ 12 ]. This is even more effective if the correlation with diseases is “mediated” by the molecules they interact with. Based on this observation, we have designed three novel prediction methods that all consider the notion of lncRNA “neighbors”, intended as lncRNAs which share common mediators among the molecules they physically interact with. Here, we focus on miRNAs as mediator molecules. However, the proposed approaches are general enough to allow also the inclusion of other different molecules. Relationships among lncRNAs, mediators and diseases are modeled through tripartite graphs in all the proposed approaches (see Fig. 1 that illustrates the flowchart of the presented research pipeline).
Problem statement Let \({\mathcal {L}}=\{l_1, l_2, \ldots , l_h\}\) be a set of lncRNAs and \({\mathcal {D}}=\{d_1, d_2, \ldots , d_k\}\) be a set of diseases. The goal is to return an ordered set of triplets \({\mathcal {R}}=\{\langle l_x, d_y, s_{xy}\rangle \}\) (with \(x\in [1,h]\) , and \(y\in [1,k]\) ), ranked according to the score \(s_{xy}\) .
The top triplets in \({\mathcal {R}}\) correspond to those pairs \((l_x, d_y)\) with most chances to represent putative LDAs which may be considered for further analysis in laboratory, while the triplets in the bottom correspond to lncRNAs and diseases which are unlikely to be related each other. A key aspect for the solution of the problem defined above is the score computation, that is the main aim of the approaches introduced in the following.
NGH: neighborhood based approach
A model of tripartite graph is adopted here to take into account that lncRNAs interacting with common mediators may be involved in common diseases.
Let \(T_{LMD}=\langle I, A \rangle\) be a tripartite graph defined on the three sets of disjoint vertexes L , M and D , such that \((l,m) \in I\) are edges between vertexes \(l \in L\) and \(m \in M\) , \((m,d) \in A\) are edges between vertexes \(m \in M\) and \(d \in D\) , respectively. In particular, L is associated to a set of lncRNAs, M to a set of miRNA and D to a set of diseases. Moreover, edges of the type ( l , m ) represent molecular interactions between lncRNAs and miRNA, experimentally validated in laboratory; edges of the type ( m , d ) correspond to known miRNA-disease associations, according to the existing literature. In both cases, interactions and associations annotated and stored in public databases may be taken into account.
The following definitions hold.
Definition 1
(Neighbors) Two lncRNAs \(l_h, l_k \in L\) are neighbors in \(T_{LMD}=\langle I, A \rangle\) if there exists at least a \(m_x \in M\) such that \((l_h, m_x) \in I\) and \((l_k, m_x) \in I\) .
Definition 2
(Prediction Score) The Prediction Score for the pair \((l_i,d_j)\) such that \(l_i \in L\) and \(d_j \in D\) is defined as:
\(M_{l_i}\) is the set of annotated miRNA interacting with \(l_i\) ,
\(M_{d_j}\) is the set of miRNA found to be associated to \(d_j\) ,
\(M_{l_x}\) is the set of miRNA interacting with the neighbor \(l_x\) of \(l_i\) (for each neighbor of \(l_i\) ),
\(\alpha\) is a real value in [0, 1] used to balance the two terms of the formula.
Definition 3
(Normalized prediction score) The Normalized Prediction Score for the pair \((l_i,d_j)\) such that \(l_i \in L\) , \(d_j \in D\) and \(s_{ij}\) is the Prediction Score for \((l_i,d_j)\) , is defined as:
NGH-CF: NGH extended with collaborative filtering
We remark that the main idea here is trying to infer the behaviour of a lncRNA, from that of its neighbors. Moreover, it is worth to point out that the notion of neighbor is related to the presence of miRNAs interacting with the same lncRNAs. However, not all the miRNA-lncRNA interactions have already been discovered, and miRNA-disease associations as well. This intuitively reminds to a typical context of data incompleteness where Collaborative Filtering may be successful in supporting the prediction process [ 30 ].
In more detail, what to be encoded by the Collaborative Filter is that lncRNAs presenting similar behaviours in terms of interactions with miRNAs, should reflect such a similarity also in their involvement with the occurrence and progress of diseases, mediated by those miRNAs. To this aim, a matrix R is considered here such that each element \(r_{ij}\) represents if (or to what extent) the lncRNA i and the disease j may be considered related. We call R relationship matrix (it is also known as rating matrix in other contexts, such as for example in the prediction of user-item associations). How to obtain \(r_{ij}\) is at the basis of the two variants of the approach presented in this section.
Due to the fact that R is usually a very sparse matrix, it can be factored into other two matrices L and D such that R \(\approx\) \(L\) \(^T\) \(D\) . In particular, matrix factorization models map both lncRNAs and diseases to a joint latent factor space F of dimensionality f , such that each lncRNA i is associated with a vector \(l_i \in F\) , each disease j with a vector \(d_j \in F\) , and their relationships are modeled as inner products in that space. Indeed, for each lncRNA i , the elements of \(l_i\) measure the extent to which it possesses those latent factors, and the same holds for each disease j and the corresponding elements of \(d_j\) . The resulting dot product in the factor space captures the affinity between lncRNA i and disease j , with reference to the considered latent factors. To this aim, there are two important tasks to be solved:
Mapping lncRNAs and diseases into the corresponding latent factors vectors.
Fill the matrix R , that is, the training set.
To learn the factor vectors \(l_i\) and \(d_j\) , a possible choice is to minimize the regularized squared error on the set of known relationships:
where \(\chi\) is the set of ( i , j ) pairs for which \(r_{ij}\) is not equal to zero in the matrix R . To this aim, we apply the ALS technique [ 31 ], which rotates between fixing the \(l_i\) ’s and fixing the \(d_j\) ’s. When all \(l_i\) ’s are fixed, the system recomputes the \(d_j\) ’s by solving a least-squares problem, and vice versa.
Filling the matrix R is performed according to two different criteria, resulting in the two different variants of the approach presented in this section, namely, CF and NGH-CF, respectively. According to the first criteria (CF), \(r_{ij}\) is set equal to 1 if the lncRNA i and the disease j share at least one miRNA in common, to 0 otherwise. The second variant (NGH-CF) works instead as a booster to improve the accuracy of NGH. In this latter case, the matrix R is filled by the normalized score ( 2 ). For both variants, the considered score to rank the predicted LDAs is given by the final value returned by the ALS technique applied on the corresponding matrix R .
Validation methodologies
We remark that the proposed approaches for LDAs prediction return a rank of LDAs, sorted according to the score that is characteristic of the considered approach, such that top triplets may be assumed as the most promising putative LDAs for further analysis in laboratory. As in other contexts [ 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 ], the performance of a prediction tool may be evaluated using suitable external criteria . Here, an external criterion relies on the existence of LDAs that are known to be true from the literature or, even better, from public repositories, where associations already verified in laboratory are annotated. A gold standard is constructed, containing only such true LDAs. The putative LDAs returned by the prediction method can thus be compared against those in the gold standard. In order to work properly, this validation methodology requires the gold standard information to be independent on that considered, in its turn, from the method under evaluation during its prediction task. This is satisfied in our case, due to the fact that all three approaches introduced in the previous sections do not exploit any type of knowledge referred to known LDAs during prediction, relying instead on known miRNA-lncRNA interactions and miRNA-disease associations, which come from independent sources.
According to the above mentioned validation methodology, the proposed approaches can be validated with references to the Receiver Operating Characteristics (ROC) analysis [ 34 ]. In particular, each predicted LDA is associated to a label, that is true if that association is contained in the considered gold standard, and false otherwise.
By varying the threshold value, it is possible to compute the true positive rate (TPR) and the false positive rate (FPR), by refferring to the percentage of the true/false predictions whose ranking is higher/below than the considered threshold value. ROC curve can be drawn by plotting TPR versus FPR at different threshold values. The Area Under ROC Curve (ROC-AUC) is further calculated to evaluate the performance of the tested methods. ROC-AUC equal to 1 indicates perfect performance, ROC-AUC equal to 0.5 random performance.
Similarly to the ROC curve, the Precision-Recall (PR) curve can be drawn as well, combining the positive predictive value (PPV, Precision), i.e., the fraction of predicted LDAs which are true in the gold standard, and the TPR (Recall), in a single visualization, at the threshold varying. The higher on y-axis the obtained curve is, the better the prediction method performance. The Area Under PR curve (AUPR) is more sensitive than AUC to the improvements for the positive class prediction [ 35 ], that is important for the case studied here. Indeed, only true LDAs are known, therefore no negative samples are included in the gold standard.
Another important measure useful to evaluate the prediction accuracy of a method and that can be considered here is the F1-score, defined as the harmonic mean of Precision and Recall to symmetrically represent both metrics in a single one.
We have validated the proposed approaches on both syntetic and real datasets, as explained below.
Synthetic data
A synthetic dataset has been built with 15 lncRNAs, 35 miRNA and 10 diseases, such that three different sets of LDAs may be identified, as follows (see also Table 1 , where the characteristics of each LDA are summarized).
Set 1: 26 LDAs, such that each lncRNA has from 3 to 4 miRNAs shared with the same disease (strongly linked lncRNAs) .
Set 2: 16 LDAs, each lncRNA having only one miRNA shared with a disease, and from 2 to 5 neighbors that are strongly linked with that same disease (directly linked lncRNAs and strong neighborhood) .
Set 3: 12 LDAs involving lncRNAs without any miRNA in common with a certain disease, and a number between 2 and 5 neighbors that are strongly linked with that same disease (only strong neighborhood) .
Experimentally verified data downloaded from starBase [ 36 ] and from HMDD [ 37 ] have been considered for the lncRNA-miRNA interactions and for the miRNA-disease associations, respectively. In particular, the latest version of HMDD, updated at 2019, has been used. Overall, \(1,\!114\) lncRNAs, \(1,\!058\) miRNAs, 885 diseases, \(10,\!112\) lncRNA-miRNA interactions and \(16,\!904\) miRNA-disease associations have been included in the analysis.
In order to evaluate the prediction accuracy of the approaches proposed here against those from the literature, three different gold standards have been considered. A first gold standard dataset GS1 has been obtained from the LncRNA-Disease database [ 38 ], resulting in 183 known and verified LDAs. A second, more restrictive, gold standard GS2 with 157 LDAs has been built by the intersection of data from [ 38 ] and [ 39 ]. Finally, also a larger gold standard dataset GS3 has been included in the analysis, by extracting LDAs from MNDRv2.0 database [ 40 ], where associations both experimentally verified and retrieved from manual literature curation are stored, resulting in 408 known LDAs.
Comparison on real data
The approaches proposed here have been compared against other approaches from the literature, over the three different gold standards described in the previous Section. In particular, all approaches considered from the literature have been run according to the default setting of their parameters, reported on the corresponding scientific publications and/or on their manual instructions.
Our approaches have been compared at first on GS1 against those approaches taking exactly the same input than ours, that are HGLDA [ 11 ], ncPred [ 10 ] and LDA-TG [ 24 ]. In particular, we have implemented HGLDA and used the corresponding p-value score, corrected by FDR as suggested by [ 11 ], for the ROC analysis. Moreover, we have normalized also the scores returned by ncPred and LDA-TG for the predicted LDAs, according to the formula in Definition 3 . Indeed, we have observed experimentally that such a normalization improves the accuracy of both methods from the literature, resulting in a better AUC. As for the novel approaches proposed here, the Normalized Prediction Score has been considered for NGH, while the approximated rating score resulting from ALS [ 31 ] is used for both CF and NGH-CF. Figure 2 shows the AUC scored by each method on GS1, while in Fig. 3 the different ROC curves are plotted. In particular, NGH scores a value of AUC equal to 0.914, thus outperforming the other three methods previously presented in the literature, i.e., HGLDA, ncPred and LDA-TG, that reach 0.876, 0.886 and 0.866, respectively (we remark also that performance of both ncPred and LDA-TG has been slightly improved with respect to their original one, by normalizing their scores). As for the novel approaches based on collaborative filtering, they both present a better accuracy than the others, with CF having AUC equal to 0.957 and NGH-CF to 0.966, respectively. Therefore, these results confirm that taking into account the collaborative effects of lncRNAs and miRNAs is useful to improve LDAs prediction, and the most successful approach is NGH-CF, that is, the neighborhood based approach boosted by collaborative filtering.
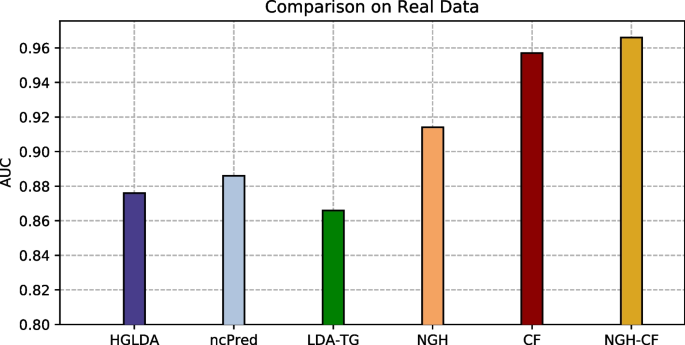
Comparison of the scored AUC on GS1
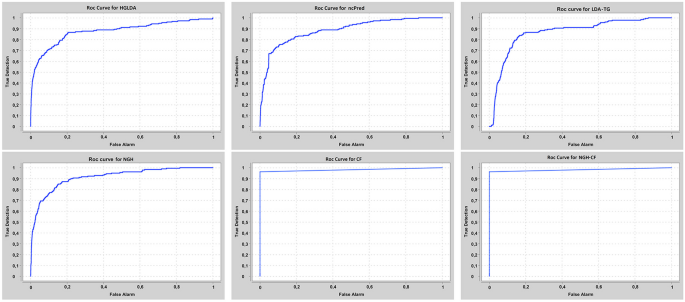
ROC curves for the compared methods on GS1
Another interesting issue is represented by the “agreement” between the different methods taking the same input, in terms of the returned best scoring LDAs. Table 2 shows the Jaccard Index computed between the proposed approaches and those receiving the same input, on the top \(5\%\) LDAs in the corresponding ranks, sorted from the best to the worst score values for each method. It emerges that results by HGLDA and ncPred have a small match with the other approaches (at most 0.23), while NGH-CF has high agreement with CF (0.74), as well as with NGH and LDA-TG (both 0.70). LDA-TG and CF present a sufficient match in their best predictions (0.59). This latter comparison based on agreement shows that approaches based on neighborhood analysis share a larger set of LDAs, in the top part of their ranks.
The proposed approaches have been compared also against other two recent methods from the literature, i.e., SIMCLDA and HGNNLDA, which receive in input different data than ours, including mRNA and drugs. For this reason, the more restrictive gold standard GS2 has been exploited for the comparison, where only lncRNAs and diseases having some correspondences with the additional input data of SIMCLDA and HGNNLDA are included. Figure 4 shows the comparison of the scored AUC on GS2, while Fig. 5 the corresponding ROC curves. In particular, the behaviour of all approaches previously tested does not change significantly on this other gold standard, moreover all the other approaches overcome SIMCLDA. On the other hand, HGNNLDA has a better performance than HGLDA, NcPred and LDA-TG, although it has a worse accuracy than NGH, CF and NGH-CF. The former confirms its superiority with regards to all considered approaches.
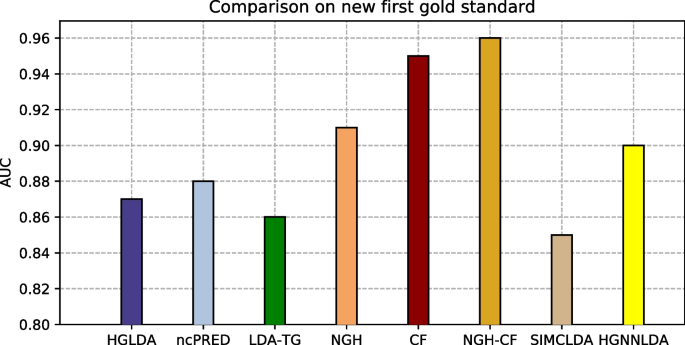
Comparison of the scored AUC on GS2
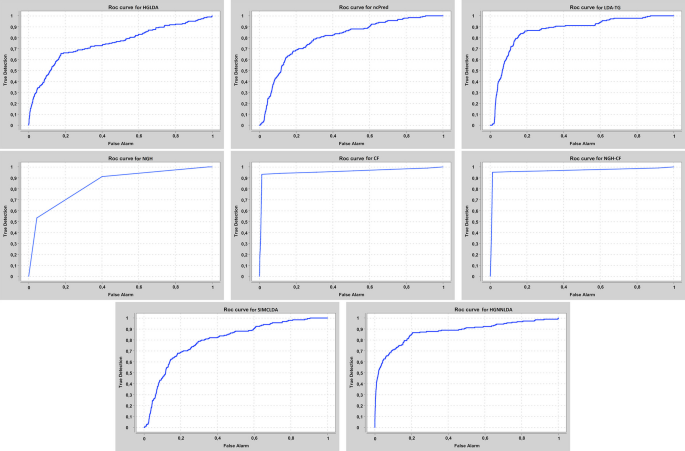
ROC curves for the compared methods on GS2
The proposed approaches have been compared also against LDAP-WMPS on GS3. Figure 6 shows the AUC values scored by all compared approaches on GS3, while Fig. 7 the corresponding ROC curves. In particular, the behaviour of all approaches previously tested does not change on this other gold standard, and LDAP-WMPS has better performance than the other approaches except for NGH, CF, NGH-CF and HGNNLDA.
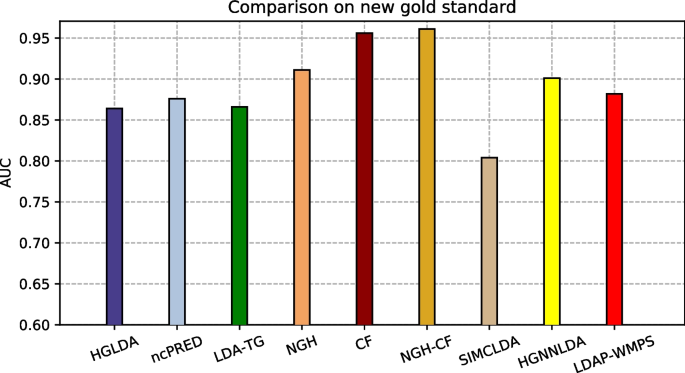
Comparison of the scored AUC on GS3
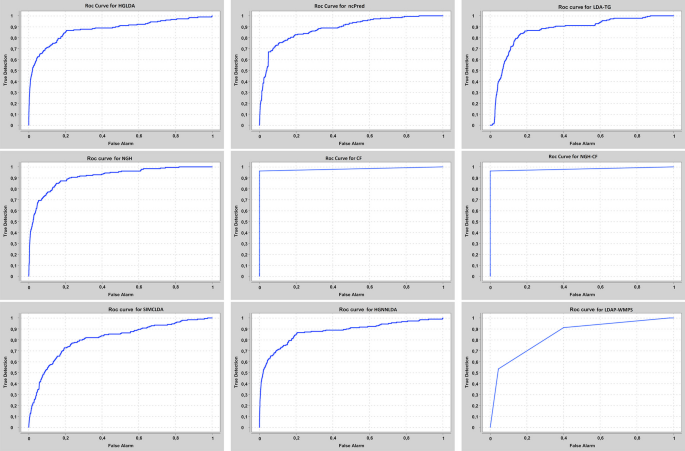
ROC curves for the compared methods on GS3
The AUPR values scored by the compared methods on GS1, GS2, and GS3 are shown in Fig. 8 , while the corresponding PR-curves are plotted in Fig. 9 . In particular, for GS1 results are analogous to the ROC analysis, with NGH-CF the best performing one, followed by CF and NGH, while HGLDA is the worst. On GS2, NGH-CF and CF keep their superiority, followed by SMCLDA and NGH, while HGLDA is yet the worst one. On GS3, NGH-CF is the first, Cf the second and both HGNNLDA and LDAP-WMPS outperform NGH, while HGLDA in this case slightly outperforms LDA-TG, ncPred and SMCLDA, which results to be the worst one.
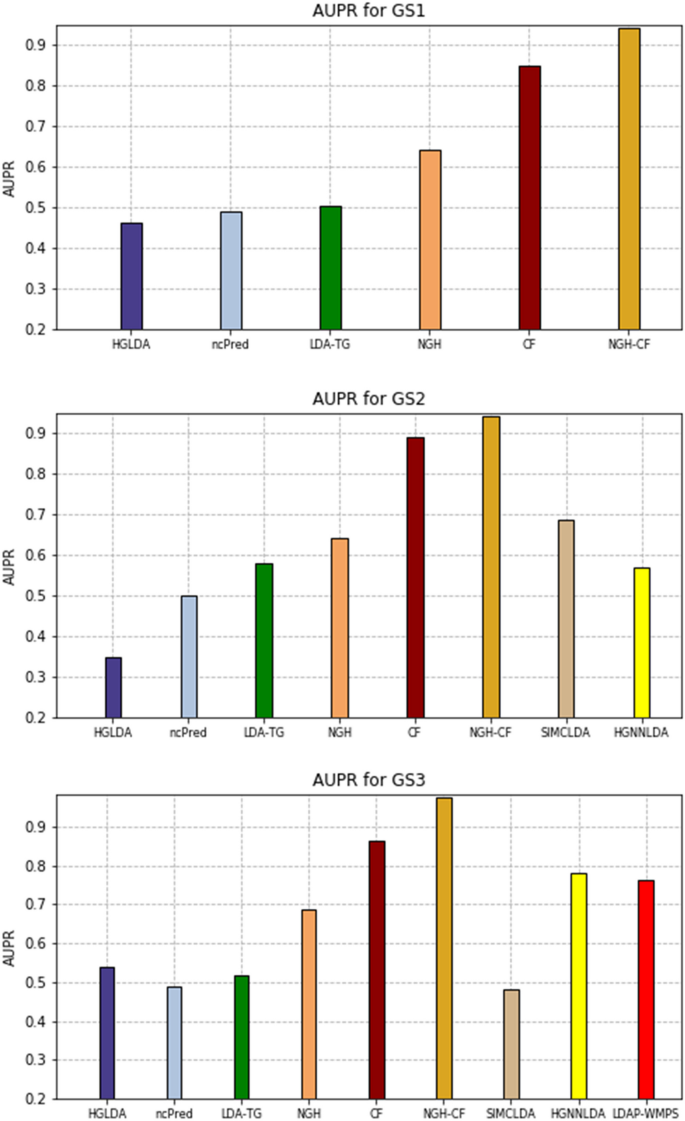
AUPR hystogram for the compared methods on GS1, GS2, GS3
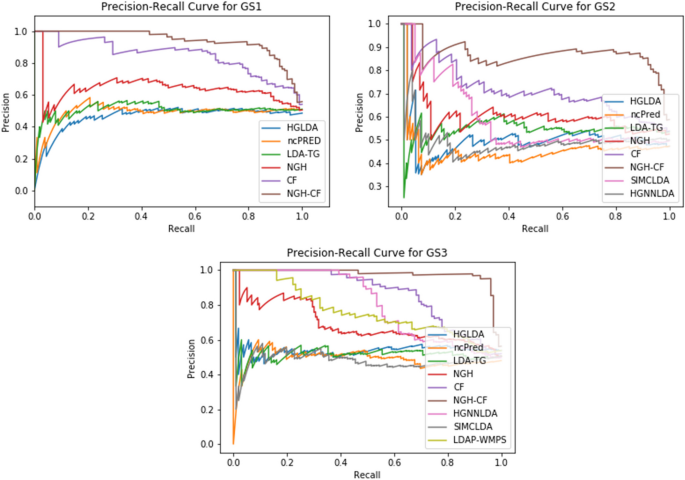
Precision-recall curves for the compared methods on GS1,GS2,GS3
Figures 10 , 11 and 12 show the F1-score values obtained, for all methods compared on GS1, GS2 and GS3, respectively, at the varying of a threshold fixed on the method score. In Tables 3 , 4 and 5 it is shown, for each gold standard, the highest value of F1-score obtained by each considered method, as well as the corresponding Precision and Recall values, and the minimum threshold value for which the highest F1-score value has been reached. On GS1 and GS2, the three best performing approaches are NGH-CF, CF and NGH, in this order. On GS3 the order is the same, and LDAP-WMPS performs equally to NGH.
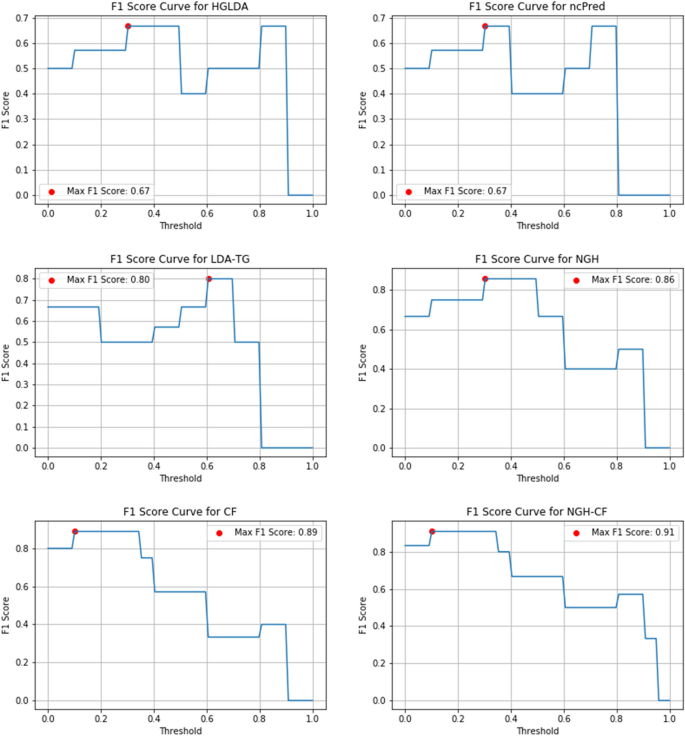
F1-score for the compared methods on GS1
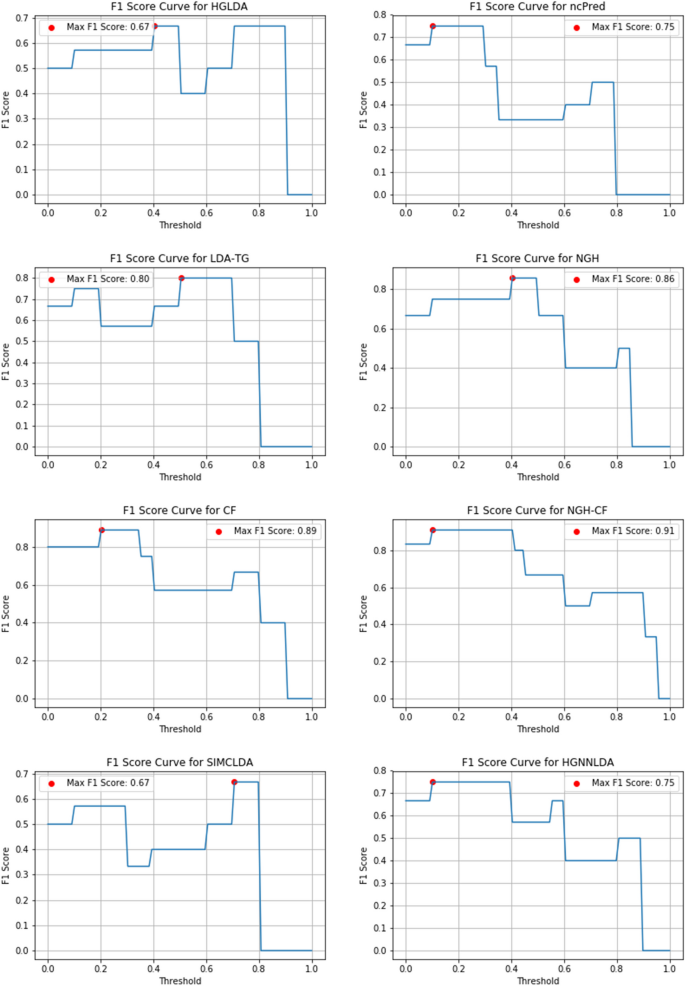
F1-Score for the compared methods on GS2
F1-Score for the compared methods on GS3
Robustness analysis
The main aim of the analysis discussed here is to measure to what extent the proposed methods are able to correctly recognize verified LDAs, even if part of the existing associations are missed, i.e., the sets of known and verified lncRNA-miRNA interactions and miRNA-disease associations are not complete. This is important to verify that the proposed approaches can provide reliable predictions also in presence of data incompleteness, that is often the case when lncRNAs are involved. Therefore, the robustness of each proposed method has been evaluated by performing progressive alterations of the input associations coming from the real datasets, according to the following three different criteria.
Progressively eliminate the \(5\%\) , \(10\%\) , \(15\%\) and \(20\%\) of lncRNA-miRNA interactions from the input data.
Progressively eliminate the \(5\%\) , \(10\%\) , \(15\%\) and \(20\%\) of miRNA-disease associations from the input data.
Progressively eliminate the \(5\%\) , \(10\%\) , \(15\%\) and \(20\%\) of both lncRNA-miRNA interactions and miRNA-disease associations (half and half), from the input data.
Tests summarized above have been performed for 20 times each. Tables 6 , 7 and 8 show the mean of the AUC values for NGH, CF and NGH-CF, respectively, over the 20 tests. In particular, all methods perform well on the three test typologies at \(5\%\) , the worst being NGH-CF, which however presents an average AUC equal to 0.84 for case 1), that is still a high value. NGH-CF is also the method that presents the best robustness on case 3), keeping the value of 0.92 also at \(20\%\) , while CF is the worst performing in case 3), indeed its average AUC decreases from 0.95 at \(5\%\) to 0.63 already at \(10\%\) , and then to 0.50 at \(20\%\) . This behaviour in case 3), where both lncRNA-miRNA interactions and miRNA-disease associations are progressively eliminated, deserves some observations. Indeed, results show that the combination of neighborhood analysis and collaborative filtering is the most robust one with regards to this perturbation, while collaborative filtering alone is the worst performing. On the other hand, CF results to be the most robust in case 1), where only lncRNA-miRNA interactions are eliminated, and this is due to the fact that CF does not take into account how many miRNAs are shared by pairs of lncRNAs. As for case 2), performance of all methods is comparable and generally good, possibly in consideration of the fact that a large number of miRNA-disease associations are available, therefore discarding small percentages of them does not affect largely the final prediction.
Comparison on specific situations
In this section further experimental tests are described, showing how well the considered methods perform in detecting specific situations, depicted through the synthetic dataset first, and then searched for in the real data. In particular, the basic observation here is that prediction approaches from the literature usually fail in detecting true LDAs, when the involved lncRNAs and diseases do not have a large number of shared miRNAs (referring to those approaches taking the same input than ours). The novel approaches we propose are particularly effective in managing the situation depicted above, through neighborhood analysis and collaborative filtering, allowing to detect similar behaviours shared by different lncRNAs, depending on the miRNAs they interact with.
For each set of LDAs defined in the synthetic data (i.e., set 1, set 2, and set 3), and for each tested method (i.e., HGLDA, NCPRED, NHG, CF, NGH-CF), Table 9 shows the percentage of LDAs in that set which is recognized at the top \(10\%\) , \(20\%\) , \(30\%\) , \(50\%\) of the rank of all LDAs, sorted by the score returned by the considered method. As an example, for HGLDA the \(32\%\) of LDAs of set 1 are located in the top \(10\%\) of its rank, where instead none LDAs in set 2 or 3 find place.
Looking at these results some interesting considerations come out. First of all, for the methods HGLDA, NCPRED, NHG and CF most associations of the set 1 are located in the top \(50\%\) of their corresponding ranks, while NGH-CF has a different behaviour. Indeed, it locates a lower number of such LDAs in the highest part of its rank than the other approaches, possibly due to the fact that it leaves room for a larger number of associations in the other two sets in the top ranked positions. As for LDAs in the set 2, all methods recognize some of them already in the top \(10\%\) , except for HGLDA, as alredy highlighted. The approaches able to recognize the larger percentages of these associations at the top \(50\%\) of their rank are NGH and NGH-CF. LDAs in the set 3 are the most difficult to recognize, due to the fact that the lncRNA and the disease do not share any miRNA in common. Indeed, the worst performing methods in this case are HGLDA, which is able to locate some of these associations only at the top \(50\%\) (according to the percentages we considered here), and NCPRED, which performs slightly better although it reaches the same percentage of located associations than HGLDA at \(50\%\) (the \(28\%\) ). As expected, approaches based on neighborhood analysis and collaborative filtering perform better, with the best one resulting to be NGH-CF.
In the previous section we have shown that all methods proposed here are able to detect specific situations, characterized by the fact that a lncRNA may have very few (or none) common miRNAs with a disease, and its neighbors share instead a large set of miRNAs with that disease. We have checked if this case occurs among the verified LDAs that our approaches find and their competitors do not. Table 10 shows, only by meaning of example, 10 experimentally verified LDAs, included in GS1, that are top ranked for the novel approaches proposed here, whereas they are in the bottom rank of the other approaches from the literature compared on GS1. Six out of such LDAs do not present any common miRNAs between the lncRNA and the disease, while four share only one miRNA. All involved lncRNAs present neighbors with a large number of miRNAs in common with the disease in that LDA, in accordance with the hypothesis that the ability in capturing this situation allows to obtain a better accuracy.
Survival analysis has been also performed by one of the TCGA Computational Tools, that is, TANRIC [ 41 ], on four of the pairs in Table 10 . In particular, those lncRNAs and diseases available in TANRIC have been chosen. Results are reported in Figures 13 , 14 , 15 and 16 , showing that the over-expression of the considered lncRNA determines a lower survival probability over the time, for all four considered cases.
Survival analysis related to SNHG16 and bladder neoplasm
Survival analysis related to CBR3-AS1 and prostate neoplasm
Survival analysis related to MALAT1 and bladder neoplasm
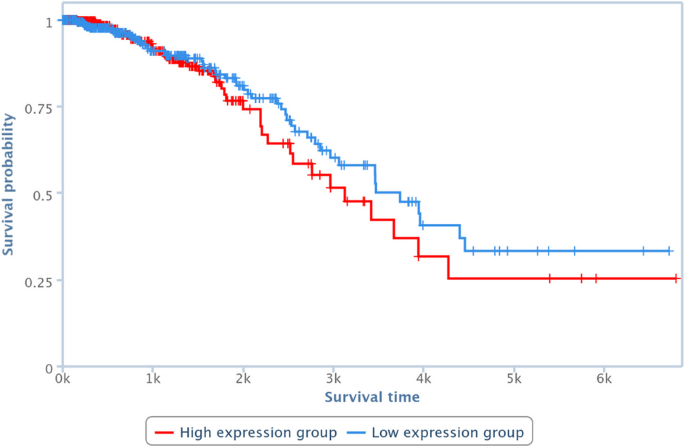
Survival analysis related to MEG3 and breast neoplasm
In the previous sections the effectiveness and robustness of the proposed approaches have been illustrated, showing that all three are able to return reliable predictions, as well as to detect specific situations which may occur in true predictions and are missed by competitors. Here we provide a discussion on some novel LDAs predicted by NGH, CF and NGH-CF.
Table 11 shows seven LDAs which are not present in the considered gold standards, and that have been returned by all three methods proposed here, with highest score. The first of these associations is between CDKN2B-AS1 and LEUKEMIA, confirmed by recent literature [ 42 , 43 ]. Indeed, CDKN2B-AS1 was found to be highly expressed in pediatric T-ALL peripheral blood mononuclear cells [ 42 ], moreover genome-wide association studies show that it is associated to Chronic Lymphocytic Leukaemia risk in Europeans [ 43 ]. As for the second association between DLEU2 and LEUKEMIA, DLEU2 is a long non-coding transcript with several splice variants, which has been identified by [ 44 ] through a comprehensive sequencing of a commonly deleted region in leukemia (i.e., the 13q14 region). Different investigations reported up regulation of this lncRNA in several types of cancers. The lncRNA H19 regulates GLIOMA angiogenesis [ 45 , 46 ], while MAP3K14 is one of the well-recognized biomarkers in the prognosis of renal cancer, which is reminiscent of the pancreatic metastasis from renal cell carcinoma [ 47 ]. MEG3 has been recently found to be important for the prediction of LEUKEMIA risk [ 48 ]. Multiple studies have shown that MIR155HG is highly expressed in diffuse large B-cell (DLBC) lymphoma and primary mediastinal B-cell lymphoma, and in chronic lymphocytic leukemia. The transcription factor MYB activates MIR155HG activity, which causes the epigenetic state of MIR155HG to be dysregulated and causes an abnormal increase in MIR155 [ 49 ]. Also the last top-ranked association in Table 11 between TUG1 and NON-SMALL CELL LUNG CARCINOMA has found evidence in the literature [ 50 , 51 , 52 ].
Tables 12 , 13 , and 14 show the top 100 (sorted by the scores returned by each method) novel LDA predictions that NGH and CF, NGH and NGH-CF, CF and NGH-CF have in common, respectively. Many of the lncRNAs involved in such top-ranked LDAs are not yet characterized in the literature, therefore results presented here may be considered a first attempt to provide novel knowledge about them, through their inferred association with known diseases.
We have explored the application of neighborhood analysis, combined with collaborative filtering, for the improvement of LDAs prediction accuracy. The three approaches proposed here have been evaluated and compared first against their direct competitors from the literature, i.e., the other methods which also use lncRNA-miRNA interactions and miRNA-disease associations, without exploiting a priori known LDAs. It results that all methods proposed here are able to outperform direct competitors, the best one (NGH-CF) also significantly (AUC equal to 0.966 against the 0.886 by NCPRED). In particular, it has been shown that the improvement in accuracy is due to the fact that our approaches capture specific situations neglected by competitors, relying on similar lncRNAs behaviour in terms of their interactions with the considered intermediate molecules (i.e., miRNAs). The proposed approaches have been then compared also against other recent methods, taking different inputs (e.g., integrative approaches), and the experimental evaluation shows that they are able to outperform them as well.
It is worth pointing out the importance of providing reliable data in input to the LDAs prediction approaches. As discussed in this manuscript, information on the lncRNAs relationships with other molecules, and between intermediate molecules and diseases, is provided in input to the proposed approaches. Reliable datasets have been used to perform the experimental analysis provided here. However, as the user may provide also different input datasets, it is important to point out that the reliability of the obtained predictions strictly depends on that of input information.
As neighborhood analysis has resulted to be effective in characterizing lncRNAs with regards to their association with known diseases, we plan to apply it also for predicting possible common functions among lncRNAs, for example by clustering them according to their interactions, which has shown to be successful for other types of molecules [ 53 ]. Moreover, due to the success of integrative approaches on the analysis of biological data [ 54 ], we expect that including other types of intermediate molecules, such as for example genes and proteins, in the main pipeline of the proposed approaches may further improve their accuracy.
In conclusion, the use of reliable input data and the integration of different types of information coming from molecular interactions seem to be the most promising future directions for LDAs prediction.
Availability of data and materials
The source code is available at: https://github.com/marybonomo/LDAsPredictionApproaches.git In particular, executable software for NGH, CF, and NGH-CF are provided, as well as syntetic and real input datasets used here; the three different gold standard datasets GS1, GS2, GS3; the final obtained results.
Medico-Salsench E, et al. The non-coding genome in genetic brain disorders: New targets for therapy? Essays Biochem. 2021;65(4):671–83.
Article CAS PubMed PubMed Central Google Scholar
Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118.
Article CAS PubMed Google Scholar
Zhao H, Shi J, Zhang Y, et al. LncTarD: a manually-curated database of experimentally-supported functional lncRNA–target regulations in human diseases. Nucl Acids Res. 2019;48(D1):D118–D126. ISSN: 0305-1048.
Liao Q, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co- expression network. Nuc Acids Res. 2011;39:3864–78.
Article CAS Google Scholar
Chen X, et al. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinf. 2017;18(4):558–76.
CAS Google Scholar
Wang B, et al. lncRNA-disease association prediction based on matrix decomposition of elastic network and collaborative filtering. Sci Rep. 2022;12:7.
Google Scholar
He J, et al. HOPEXGB: a consensual model for predicting miRNA/lncRNA-disease associations using a heterogeneous disease-miRNA-lncRNA information network. J Chem Inf Model 2023
Zhong H, et al. Association filtering and generative adversarial networks for predicting lncRNA-associated disease. BMC Bioinf. 2023;24(1):234.
Dengju Y, et al. GCNFORMER: graph convolutional network and transformer for predicting lncRNA-disease associations. BMC Bioinf. 2024;25(1):5.
Article Google Scholar
Alaimo S, Giugno R, Pulvirenti A. ncPred: ncRNA-disease association prediction through Tripartite network-based inference. Front Bioeng Biot. 2014;2:71.
Chen X. Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci Rep. 2015;5:13186.
Lu C, et al. Prediction of lncRNA-disease associations based on inductive matrix completion. Bioinformatics. 2018;34(19):3357–64.
Xuan Z, Li J, Yu X, Feng J, et al. A probabilistic matrix factorization method for identifying lncRNA-disease associations. Genes 2019;10(2)
Du X, et al. lncRNA-disease association prediction method based on the nearest neighbor matrix completion model. Sci Rep. 2022;12(1):21653.
Wang L, et al. Prediction of lncRNA-disease association based on a Laplace normalized random walk with restart algorithm on heterogeneous networks. BMC Bioinf. 2022;23(1):1–20.
Huang L, Zhang L, Chen X. Updated review of advances in microRNAs and complex diseases: taxonomy, trends and challenges of computational models. Brief Bioinf. 2022;23(5):bbac358.
Huang L, Zhang L, Chen X. Updated review of advances in microRNAs and complex diseases: experimental results, databases, webservers and data fusion. Brief Bioinf. 2022;23(6):bbac397.
Huang L, Zhang L, Chen X. Updated review of advances in microRNAs and complex diseases: towards systematic evaluation of computational models. Brief Bioinf. 2022;23(6):bbac407.
Chen X, Yan G. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics. 2013;29(20):2617–24.
Xie G, et al. SKF-LDA: similarity kernel fusion for predicting lncRNA-disease association. Mol Therapy-Nucleic Acids. 2019;18:45–55.
Liu D, et al. HGNNLDA: predicting lncRNA-drug sensitivity associations via a dual channel hypergraph neural network. IEEE/ACM transactions on computational biology and bioinformatics, 2023;1–11.
Zhang Y, et al. LDAI-ISPS: lncRNA-disease associations inference based on integrated space projection scores. Int J Molecular Sci. 2020;21(4):1508.
Liang Y, et al. MAGCNSE: predicting lncRNA-disease associations using multi-view attention graph convolutional network and stacking ensemble model. BMC Bioinf. 2022;23(1):189.
Bonomo M, La Placa A, Rombo SE. Prediction of lncRNA-disease associations from tripartite graphs. In: Heterogeneous data management, polystores, and analytics for healthcare - VLDB workshops, poly 2020 and DMAH 2020, virtual event, August 31 and September 4, 2020, Revised Selected Papers. Springer, Berlin, 2020;205–210. ISSN: 978-3-030-71054-5
Xie G, et al. Predicting lncRNA-disease associations based on combining selective similarity matrix fusion and bidirectional linear neighborhood label propagation. Brief Bioinform. 2023;24(1):bbac595.
Article PubMed Google Scholar
Cheng L, et al. ntNetLncSim: an integrative network analysis method to infer human lncRNA functional similarity. Oncotarget. 2016;7(30):47864–74.
Article PubMed PubMed Central Google Scholar
Guangyuan F, et al. Matrix factorization-based data fusion for the prediction of lncRNA-disease associations. Bioinformatics. 2018;34:1529–37.
Xie G, et al. RWSF-BLP: a novel lncRNA-disease association prediction model using random walk-based multi-similarity fusion and bidirectional label propagation. Mol Genet Genom. 2021;296:473–83.
Wang B, et al. lncRNA-disease association prediction based on the weight matrix and projection score. PLOS One. 2023;18(1): e0278817.
Duan R, Jiang C, Jain HK. Combining review-based collaborative filtering and matrix factorization: a solution to rating’s sparsity problem”. Decis Support Syst 2022;156:113748. ISSN: 0167–9236.
Koren Y, Bell R, Volinsky C. Matrix factorization techniques for recommender systems. Computer. 2009;42(8):30–7.
Parida L, Pizzi C, Rombo SE. Irredundant tandem motifs. Theoret Comput Sci. 2014;525:89–102.
Bonomo M, et al. Topological ranks reveal functional knowledge encoded in biological networks: a comparative analysis. Brief Bioinform. 2022;23(3):bbac101.
Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27(8):861–74.
Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLOS One. 2015;10(3): e0118432.
Li J, et al. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2013;42:D92–7.
Li Y, et al. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014;42:D1070–4.
Chen G, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–6.
Gao Y, et al. Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021;49(D1):D1251–8.
Cui T, et al. MNDR v2. 0: an updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 2018;46(D1):D371–4.
CAS PubMed Google Scholar
Li J, et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75(18):3728–37.
Chen L, et al. lncRNA CDKN2B-AS1 contributes to tumorigenesis and chemoresistance in pediatric T-cell acute lymphoblastic leukemia through miR-335-3p/TRAF5 axis. In: Anti-cancer drugs, Wolters Kluwer Health, Inc. (2020)
Song C, et al. CDKN2B-AS1: an indispensable long non-coding RNA in multiple diseases. Current Pharm Des. 2020;26(41):5335–46.
Ghafouri-Fard S, et al. Deleted in lymphocytic leukemia 2 (DLEU2): an lncRNA with dissimilar roles in different cancers. Biomed Pharmacother. 2021;133: 111093.
Jia P, et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381(2):359–69.
Liu Z, et al. LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1 α /VEGFaxis. Neoplasma. 2020;67(1):111–8.
Zhou S, et al. A novel immune-related gene prognostic Index (IRGPI) in pancreatic adenocarcinoma (PAAD) and its implications in the tumor microenvironment. Cancers. 2022;14(22):5652.
Pei J, et al. Novel contribution of long non-coding RNA MEG3 genotype to prediction of childhood leukemia risk. Cancer Genom Proteom. 2022;19(1):27–34.
Peng L, et al. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 2019;8(17):7161–73.
Zhang E, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5(5):e1243–e1243.
Lin P, et al. Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer. 2016;16:1–10.
Niu Y, et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16(1):1–13.
Pizzuti C, Rombo SE. An evolutionary restricted neighborhood search clustering approach for PPI networks. Neurocomputing. 2014;145:53–61.
Rombo SE, Ursino D (2021) Integrative bioinformatics and omics data source interoperability in the next-generation sequencing era
Download references
Acknowledgements
The authors are grateful to the Anonymous Reviewers, for the constructive and useful suggestions that allowed to significantly improve the quality of this manuscript. Some of the results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga .
PRIN “multicriteria data structures and algorithms: from compressed to learned indexes, and beyond”, Grant No. 2017WR7SHH, funded by MIUR (closed). “Modelling and analysis of big knowledge graphs for web and medical problem solving” (CUP: E55F22000270001), “Computational Approaches for Decision Support in Precision Medicine” (CUP:E53C22001930001), and “Knowledge graphs e altre rappresentazioni compatte della conoscenza per l’analisi di big data” (CUP: E53C23001670001), funded by INdAM GNCS 2022, 2023, 2024 projects, respectively. “Models and Algorithms relying on knowledge Graphs for sustainable Development goals monitoring and Accomplishment - MAGDA” (CUP: B77G24000050001), funded by the European Union under the PNRR program related to “Future Artificial Intelligence - FAIR”.
Author information
Authors and affiliations.
Kazaam Lab s.r.l., Palermo, Italy
Mariella Bonomo & Simona E. Rombo
Department of Mathematics and Computer Science, University of Palermo, Palermo, Italy
Simona E. Rombo
You can also search for this author in PubMed Google Scholar
Contributions
MB and SER equally contributed to the research presented in this manuscript. MB implemented and run the software, SER performed the analysis of results. Both authors wrote and reviewed the entire manuscript.
Corresponding author
Correspondence to Mariella Bonomo .
Ethics declarations
Ethics approval and consent to participate.
Not Applicable
Consent for publication
Competing interests.
SER is editor of BMC Bionformatics. MB has no Conflict of interest.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Bonomo, M., Rombo, S.E. Neighborhood based computational approaches for the prediction of lncRNA-disease associations. BMC Bioinformatics 25 , 187 (2024). https://doi.org/10.1186/s12859-024-05777-8
Download citation
Received : 13 December 2023
Accepted : 11 April 2024
Published : 13 May 2024
DOI : https://doi.org/10.1186/s12859-024-05777-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- LncRNA-disease associations
- Molecular interactions
- Bioinformatics
- Long non-coding RNA
BMC Bioinformatics
ISSN: 1471-2105
- General enquiries: [email protected]
Help | Advanced Search
Computer Science > Machine Learning
Title: predicting ship responses in different seaways using a generalizable force correcting machine learning method.
Abstract: A machine learning (ML) method is generalizable if it can make predictions on inputs which differ from the training dataset. For predictions of wave-induced ship responses, generalizability is an important consideration if ML methods are to be useful in design evaluations. Furthermore, the size of the training dataset has a significant impact on the practicality of a method, especially when training data is generated using high-fidelity numerical tools which are expensive. This paper considers a hybrid machine learning method which corrects the force in a low-fidelity equation of motion. The method is applied to two different case studies: the nonlinear responses of a Duffing equation subject to irregular excitation, and high-fidelity heave and pitch response data of a Fast Displacement Ship (FDS) in head seas. The generalizability of the method is determined in both cases by making predictions of the response in irregular wave conditions that differ from those in the training dataset. The influence that low-fidelity physics-based terms in the hybrid model have on generalizability is also investigated. The predictions are compared to two benchmarks: a linear physics-based model and a data-driven LSTM model. It is found that the hybrid method offers an improvement in prediction accuracy and generalizability when trained on a small dataset.
Submission history
Access paper:.
- Other Formats
References & Citations
- Google Scholar
- Semantic Scholar
BibTeX formatted citation
Bibliographic and Citation Tools
Code, data and media associated with this article, recommenders and search tools.
- Institution
arXivLabs: experimental projects with community collaborators
arXivLabs is a framework that allows collaborators to develop and share new arXiv features directly on our website.
Both individuals and organizations that work with arXivLabs have embraced and accepted our values of openness, community, excellence, and user data privacy. arXiv is committed to these values and only works with partners that adhere to them.
Have an idea for a project that will add value for arXiv's community? Learn more about arXivLabs .
Mr Salles Teaches English

An Inspector Calls Prediction 2024

What Makes a Grade 7 Essay on An Inspector Calls
Explanations 21
NAMED METHODS 3
Patriarchal 0
Thesis Statement Y
Conclusion Y
You are probably wondering what an explanation is. It is anything which means: this implies, indicates, suggests, connotes, Priestley wants us to think, feel, believe, understand..
It is easier just to show you. Explanations are in italics.
Student Essay
2022 Theme Question
Priestley has each member of the Birling family confess to their exploitation of Eva in order to reveal social inequality for his audience. He presents this as a tragedy.
The Inspector’s portrayal of Eva’s death demands a reaction from each character . While Birling is dismissive , proclaiming “I don’t see what this has to do with me” , Eric is emotional, exclaiming “My God” .
Birling is unrepentant when he explains how he sacked Eva. He describes her as a “troublemaker” who went on strike for higher wages. Priestley reveals the social inequality between businessmen and their workers by revealing the need for workers to strike .
Birling’s justification for sacking Eva is that he must “keep labour costs down” . He views her only as “cheap labour” , and Priestley thereby emphasises how businessmen viewed the working classes as commodities . Her sacking begins a “ chain of events ” culminating in tragedy. Eric describes this as a “damn shame” .
Sheila meets Eva in her new employment at Milwards. She abuses her status to force the shop to sack Eva, which emphasises social inequality through class . Sheila’s obsession with fashion and appearance is a criticism of upper class women of the time . This obsession leads her to demand Eva be sacked. Priestley USES the triviality of a dress to contrast with the tragic impact on Eva to provoke our anger at upper class exploitation of the working classes .
Gerald’s actions also emphasise this inequality and exploitation based on class . He portrays himself as heroic, hearing Eva’s “cry for help” and “rescuing” her from the sexual advances of Alderman Meggarty.
He views himself as a hero, and Eva as helpless and powerless . He exploits this weakness to keep her as his mistress, despite his heroic self-image. We are disgusted that Eva is simply a tool for his sexual gratification. We are shocked when he simply ends the relationship when it suits him. Eva must have felt exploited by him, and this must have impacted on her decision to commit suicide.
The elite status of upper class men allowed them to act in this way without punishment. For example, Birling expects to be knighted despite his exploitation. We might say he is rewarded for it rather than punished . The Inspector conveys Priestley’s viewpoint when he warns that this suffering affects “millions of Eva Smiths and John Smiths” .
Priestley also describes Mrs Birling as “her husband’s social superior” . This highlights how class divisions exist even within the family. She asserts her status by criticising her husband : “you shouldn’t say such things” . Her “cold” lack of emotion also leads to her unwittingly causing the death of her grandchild.
She rejects Eva’s appeal for financial help because of her view of class , and this leads directly to Eva’s suicide. Mrs Birling refuses to accept any responsibility, instead accusing the unborn child’s father.
In conclusion , this stubbornness and prejudice emphasises the tragedy of Eva’s death . We realise that the older generation will never change and consequently Eva’s tragedy will be replayed by many in society .
Original 615 words
Mr Salles Teaches English is a reader-supported publication. To receive new posts which help you get top grades, consider becoming a free or paid subscriber.
Examiner Comments
This answer focuses very well on the question.
It has a range of references
These are selected well to back up the argument
There are enough to make this a ‘detailed’ answer
And they are woven into the explanations to back up a range of points
This scores highly for AO1, response to task
The answer also deals with Priestley’s ideas
And the themes of the play
The comments about characters refer to Priestley’s methods in how he presents them
And the best example of this is in the discussion of Gerald
To get the top mark in Level 5, the student should analyse other characters with the same level of discussion
My Comments
Explanations 24
Thesis Statement Yes
Conclusion Yes
1. A two part thesis statement! Hurrah. I particularly like the idea of tragedy. In a Greek tragedy, characters try to avoid their fate, but a fatal flaw in their character brings the tragedy about. Does Eva have a fatal flaw, a hamartia? We would probably argue that she doesn’t, it is society which is flawed. And therefore, tragedies like Eva’s are playing out to the “millions and millions” of working class people she represents. Take this idea, and you have a grade 9 thesis statement.
2. And obviously I’m super excited about the 24 explanations. The previous answer was much shorter, with only 16 explanations. So, what is the deal?
3. Look at context. The examiner was much more excited about Priestley’s ideas about socialism and capitalism than this student’s much narrower focus on class.
4. The examiner’s comments about the analysis of Gerald are really helpful. One way of looking at this is to look at the characters from two perspectives – here it is that Gerald sees himself as heroic, but actually he is exploitative.
5. You could easily do that with Eric – he believes he is doing the right thing, standing by Eva and offering to marry her, but he has probably raped her and stolen money from his father’s business.
6. Then we have to link this to Priestley’s purpose – he might want us to question Eric’s apparent transformation, and suggest that upper class men have only a veneer of humanity about them, but privilege has fatally damaged them. They will therefore continue to exploit those around them, even those they apparently care for. You can also link this to the ending: if Eric genuinely learns the Inspector’s lesson, why is there the second phone call?
7. Mrs Birling thinks she is right to deny Eva helped because she was obviously lying (which is true) and because she believes Eva tried to insult Sybil by calling herself Mrs Birling (which may also be true). But, charity is there to help people who need it, not people we like.
8. Then we have to link this to Priestley’s purpose, which is to suggest that we need a socialist society, a welfare state, where those with power and social status are not sitting in judgement of the poor. Instead, the state will decide to act with kindness and generosity.
9. So, we’ve considered the characters from two points of view: how they see themselves, and how Priestley wants us to see them.
10. Then we have said WHY Priestley wants us to see them that way. We always relate the WHY to society and socialism. And the grade 9 falls into our laps.
Paid subscribers get a new grade 9 answer to language and literature questions every week except the summer holidays. They also access over 70 already published.
This post includes a grade 7 answer on the Inspector, and a grade 9 answer written by me to my predicted question.
Keep reading with a 7-day free trial
Subscribe to Mr Salles Teaches English to keep reading this post and get 7 days of free access to the full post archives.
- Open access
- Published: 12 May 2024
Distance plus attention for binding affinity prediction
- Julia Rahman 1 na1 ,
- M. A. Hakim Newton 2 , 3 na1 ,
- Mohammed Eunus Ali 4 &
- Abdul Sattar 2
Journal of Cheminformatics volume 16 , Article number: 52 ( 2024 ) Cite this article
200 Accesses
Metrics details
Protein-ligand binding affinity plays a pivotal role in drug development, particularly in identifying potential ligands for target disease-related proteins. Accurate affinity predictions can significantly reduce both the time and cost involved in drug development. However, highly precise affinity prediction remains a research challenge. A key to improve affinity prediction is to capture interactions between proteins and ligands effectively. Existing deep-learning-based computational approaches use 3D grids, 4D tensors, molecular graphs, or proximity-based adjacency matrices, which are either resource-intensive or do not directly represent potential interactions. In this paper, we propose atomic-level distance features and attention mechanisms to capture better specific protein-ligand interactions based on donor-acceptor relations, hydrophobicity, and \(\pi \) -stacking atoms. We argue that distances encompass both short-range direct and long-range indirect interaction effects while attention mechanisms capture levels of interaction effects. On the very well-known CASF-2016 dataset, our proposed method, named Distance plus Attention for Affinity Prediction (DAAP), significantly outperforms existing methods by achieving Correlation Coefficient (R) 0.909, Root Mean Squared Error (RMSE) 0.987, Mean Absolute Error (MAE) 0.745, Standard Deviation (SD) 0.988, and Concordance Index (CI) 0.876. The proposed method also shows substantial improvement, around 2% to 37%, on five other benchmark datasets. The program and data are publicly available on the website https://gitlab.com/mahnewton/daap.
Scientific Contribution Statement
This study innovatively introduces distance-based features to predict protein-ligand binding affinity, capitalizing on unique molecular interactions. Furthermore, the incorporation of protein sequence features of specific residues enhances the model’s proficiency in capturing intricate binding patterns. The predictive capabilities are further strengthened through the use of a deep learning architecture with attention mechanisms, and an ensemble approach, averaging the outputs of five models, is implemented to ensure robust and reliable predictions.
Introduction
Conventional drug discovery, as noted by a recent study [ 1 ], is a resource-intensive and time-consuming process that typically lasts for about 10 to 15 years and costs approximately 2.558 billion USD to bring each new drug successfully to the market. Computational approaches can expedite the drug discovery process by identifying drug molecules or ligands that have high binding affinities towards disease-related proteins and would thus form strong transient bonds to inhibit protein functions [ 2 , 3 , 4 ]. In a typical drug development pipeline, a pool of potential ligands is usually given, and the ligands exhibiting strong binding affinities are identified as the most promising drug candidates against a target protein. In essence, protein-ligand binding affinity values serve as a scoring method to narrow the search space for virtual screening [ 5 ].
Existing computational methods for protein-ligand binding affinity prediction include both traditional machine learning and deep learning-based approaches. Early methods used Kernel Partial Least Squares [ 6 ], Support Vector Regression (SVR) [ 7 ], Random Forest (RF) Regression [ 8 ], and Gradient Boosting [ 9 ]. However, just like various other domains [ 10 , 11 , 12 , 13 , 14 ], drug discovery has also seen significant recent advancements [ 15 , 16 , 17 , 18 ] from the computational power and extensive datasets used in deep learning. Deep learning models for protein-ligand binding affinity prediction take protein-ligand docked complexes as input and give binding affinity values as output. Moreover, these models use various input features to capture the global characteristics of the proteins and the ligands and their local interactions in the pocket areas where the ligands get docked into the proteins.
Recent deep learning models for protein-ligand binding affinity prediction include DeepDTA [ 19 ], Pafnucy [ 20 ], \(K_\text {DEEP}\) [ 21 ], DeepAtom [ 22 ], DeepDTAF [ 23 ], BAPA [ 5 ], SFCNN [ 24 ], DLSSAffinity [ 4 ] EGNA [ 25 ], CAPLA [ 26 ] and ResBiGAAT [ 27 ]. DeepDTA [ 19 ] introduced a Convolutional Neural Network (CNN) model with input features Simplified Molecular Input Line Entry System (SMILES) sequences for ligands and full-length protein sequences. Pafnucy and \(K_{DEEP}\) used a 3D-CNN with 4D tensor representations of the protein-ligand complexes as input features. DeepAtom employed a 3D-CNN to automatically extract binding-related atomic interaction patterns from voxelized complex structures. DeepDTAF combined global contextual features and local binding area-related features with dilated convolution to capture multiscale long-range interactions. BAPA introduced a deep neural network model for affinity prediction, featuring descriptor embeddings and an attention mechanism to capture local structural details. SFCNN employed a 3D-CNN with simplified 4D tensor features having only basic atomic type information. DLSSAffinity employed 1D-CNN with pocket-ligand structural pairs as local features and ligand SMILES and protein sequences as global features. EGNA introduced an empirical graph neural network (GNN) that utilizes graphs to represent proteins, ligands, and their interactions in the pocket areas. CAPLA [ 26 ] utilized a cross-attention mechanism within a CNN along with sequence-level input features for proteins and ligands and structural features for secondary structural elements. ResBiGAAT [ 27 ] integrates a deep Residual Bidirectional Gated Recurrent Unit (Bi-GRU) with two-sided self-attention mechanisms, utilizing both protein and ligand sequence-level features along with their physicochemical properties for efficient prediction of protein-ligand binding affinity.
In this work, we consider the effective capturing of protein-ligand interaction as a key to making further progress in binding affinity prediction. However, as we see from the literature, a sequential feature-based model such as DeepDTA was designed mainly to capture long-range interactions between proteins and ligands, not considering local interactions. CAPLA incorporates cross-attention mechanisms along with sequence-based features to indirectly encompass short-range interactions to some extent. ResBiGAAT employs a residual Bi-GRU architecture and two-sided self-attention mechanisms to capture long-term dependencies between protein and ligand molecules, utilizing SMILES representations, protein sequences, and diverse physicochemical properties for improved binding affinity prediction. On the other hand, structural feature-based models such as Pafnucy, \(K_{DEEP}\) and SFCNN use 3D grids, 4D tensors, or molecular graph representations. These features provide valuable insights into the pocket region of the protein-ligand complexes but incur significant computational costs in terms of memory and processing time. Additionally, these features have limitations in capturing long-range indirect interactions among protein-ligand pairs. DLSSAffinity aims to bridge the gap between short- and long-range interactions by considering both sequential and structural features. Moreover, DLSSAffinity uses 4D tensors for Cartesian coordinates and atom-level features to represent interactions between heavy atoms in the pocket areas of the protein-ligand complexes. These representations of interactions are still indirect, considering the importance of protein-ligand interaction in binding affinity. EGNA tried to use graphs and Boolean-valued adjacency matrices to capture protein-ligand interactions to some extent. However, EGNA’s interaction graph considers only edges between each pair of a \(C_\beta \) atom in the pocket areas of the protein and a heavy atom in the ligand when their distance is below a threshold of \(10\mathring{A}\) .
Inspired by the use of distance measures in protein structure prediction [ 14 , 28 , 29 ], in this work, we employ distance-based input features in protein-ligand binding affinity prediction. To be more specific, we use distances between donor-acceptor [ 30 ], hydrophobic [ 31 , 32 ], and \(\pi \) -stacking [ 31 , 32 ] atoms as interactions between such atoms play crucial roles in protein-ligand binding. These distance measures between various types of atoms could essentially capture more direct and more precise information about protein-ligand interactions than using sequence-based features or various other features representing the pocket areas of the protein-ligand complexes. Moreover, the distance values could more directly capture both short- and long-range interactions than adjacency-based interaction graphs of EGNA or tensor-based pocket area representations of DLSSAffinity. Besides capturing protein-ligand interactions, we also consider only those protein residues with donor, hydrophobic, and \(\pi \) -stacking atoms in this work. Considering only these selective residues is also in contrast with all other methods that use all the protein residues. For ligand representation, we use SMILES strings. After concatenating all input features, we use an attention mechanism to effectively weigh the significance of various input features. Lastly, we enhance the predictive performance of our model by adopting an ensembling approach, averaging the outputs of several trained models.
We name our proposed method as Distance plus Attention for Affinity Prediction (DAAP). On the very well-known CASF-2016 dataset, DAAP significantly outperforms existing methods by achieving the Correlation Coefficient (R) 0.909, Root Mean Squared Error (RMSE) 0.987, Mean Absolute Error (MAE) 0.745, Standard Deviation (SD) 0.988, and Concordance Index (CI) 0.876. DAAP also shows substantial improvement, ranging from 2% to 37%, on five other benchmark datasets. The program and data are publicly available on the website https://gitlab.com/mahnewton/daap .
In our study, we first demonstrate the robustness of our deep architecture through five-fold cross-validation. Subsequently, the learning curve, as depicted in Fig. 1 , illustrates the dynamics of training and validation loss, providing insights into the stability and reliability of the learning process. Furthermore, we provide a comprehensive performance comparison of our proposed model with current state-of-the-art predictors. We also provide an in-depth analysis of the experimental results. The effectiveness of our proposed features is substantiated through an ablation study and a detailed analysis of input features.
Training and validation loss curve of DAAP
Five-fold cross-validation
This study employs a five-fold cross-validation approach to evaluate the performance of the proposed model thoroughly, demonstrating the robustness of the deep architecture. Table 1 provides the average performance metrics (R, RMSE, MAE, SD, and CI) along with their corresponding standard deviations derived from the 5-fold cross-validation on the CASF \(-\) 2016.290 test set when the model is trained with PDBbind2016 and PDBbind2020 datasets. This presentation highlights the predictor’s predictive accuracy and reliability, emphasising the proposed model’s effectiveness.
Average ensemble
Our proposed approach leverages an attention-based deep learning architecture to predict binding affinity. The input feature set comprises distance matrices, sequence-based features for specific protein residues, and SMILES sequences. To enhance the robustness and mitigate the effects of variability and overfitting, we train five models and employ arithmetic averaging for ensembling. Average ensembling is more suitable than max voting ensembling when dealing with real values.
Table 2 shows the results of five models and their averages when all models have the identical setting of their training parameters and the training datasets. We see that the ensemble results are better than the results of the individual models in both the PDBbind2016 and PDBbind2020 training datasets. To check that the proposed approach is robust over the variability in the training datasets, we also train five models but each with a different training subset. These training subsets were obtained by using sampling with replacement. Table 3 shows the results of these five models and their averages.
Tables 2 and 3 depict that the ensemble results are better than the results of the individual results in both training sets. It might seem counterintuitive to see the average results are better than all the individual results, but note that these are not simple average of averages. When the ensemble results are compared across Tables 2 and 3 , the best results are observed in Table 2 for the PDBbind2020 training set. All evaluation metrics R, RMSE, SD, MAE, and CI display improved performance when using the same training data (Table 2 ) compared to different varying training data (Table 3 ) in PDBbind2020 data set. Accordingly, we choose the ensemble with the same training data for PDBbind2020 (Table 2 ) as our final binding affinity prediction model. Conversely, for PDBbind2016, superior outcomes are obtained from the varied training subsets in Table 3 . Henceforth, the best-performing models using PDBbind2016 and PDBbind2020 will be referred to as DAAP16 and DAAP20, respectively, in subsequent discussions.
Comparison with state-of-the-art methods
In our comparative analysis, we assess the performance of our proposed affinity predictor, DAAP, on the CASF-2016 test set, compared to nine recent state-of-the-art predictors: Pafnucy [ 20 ], DeepDTA [ 19 ], OnionNet [ 3 ], DeepDTAF [ 23 ], SFCNN [ 24 ] DLSSAffinity [ 4 ], EGNA [ 25 ], CAPLA [ 26 ] and ResBiGAAT [ 27 ]. Notably, the most recent predictors have surpassed the performance of the initial four, prompting us to focus our comparison on their reported results. For the latter five predictors, we detail the methodology of obtaining their results as follows:
DLSSAffinity We rely on the results available on DLSSAffinity’s GitHub repository, as direct prediction for specific target proteins is not possible due to the unavailability of its trained model.
SFCNN Utilizing the provided weights and prediction code from SFCNN, we replicate their results, except for CASF-2013. The ambiguity regarding the inclusion of CASF-2013 data in their training set (sourced from the PDBbind database version 2019) leads us to omit these from our comparison.
EGNA We have adopted EGNA’s published results for the CASF-2016 test set with 285 protein-ligand complexes due to differing Uniclust30 database versions for HHM feature construction. We applied EGNA’s code with our HHM features for the other five test sets to ensure a consistent evaluation framework.
CAPLA Predictions are made based on the features given in CAPLA’s GitHub, except for the ADS.74 dataset, where we can’t predict results due to the unavailability of feature sets. Their results are the same as their reported results.
ResBiGAAT We included ResBiGAAT’s published results in our analysis after encountering discrepancies with their online server using the same SMILES sequences and protein sequences from test PDB files as us. Variations in results, particularly for PDB files with multiple chains, led us to rely on their reported data, as it yielded more consistent and higher accuracies than our attempts.
In Table 4 , the first 8 methods, namely Pafnucy, DeepDTA, OnionNet, DeepDTAF, DLSSAffinity, SFCNN, \(EGNA^*\) and CAPLA reported on 290 CASF-2016 protein-ligand complexes. To make a fair comparison with these 8 methods, we compared our proposed method DAAP16 and DAAP20 on those 290 protein-ligand complexes. From the data presented in the Table 4 , it is clear that our DAAP20 approach outperforms all the 8 predictors, achieving the highest R-value of 0.909, the highest CI value of 0.876, the lowest RMSE of 0.987, the lowest MAE of 0.745, and the lowest SD of 0.988. Specifically, compared to the closest state-of-the-art predictor, CAPLA, our approach demonstrated significant improvements, with approximately 5% improvement in R, 12% in RMSE, 14% in MAE, 11% in SD, and 4% in CI metrics, showcasing its superior predictive capabilities. As 3 of the recent predictors, namely SFCNN, EGNA, and ResBiGAAT, reported their result for the 285 protein-ligand complexes on the CASF-2016 dataset, to make a fair comparison with them as well, we assess our predictor, DAAP, on these 285 proteins as well. From the data presented in Table 4 , the results revealed that, across all metrics, DAAP20 outperformed these three predictors on 285 proteins as well. Particularly, compared to the recent predictor ResBiGAAT, our approach demonstrated notable improvements, with around 6% improvement in R, 19% in RMSE, 20% in MAE, and 5% in CI metrics, highlighting its superior predictive capabilities.
Table 5 presents a comprehensive evaluation of the prediction performance of our proposed DAAP approach on five other well-known test sets CASF \(-\) 2013.87, CASF \(-\) 2013.195 ADS.74, CSAR-HiQ.51 and CSAR-HiQ.36. Across these test sets, our DAAP approaches demonstrate superior predictive performance in protein-ligand binding affinity. On the CASF \(-\) 2013.87 dataset, EGNA surpasses CAPLA with higher R-value and CI-value of 0.752 and 0.767, respectively, while CAPLA records lower RMSE, MAE and SD values of 1.512, 1.197, and 1.521. In contrast, our DAAP20 surpasses both, excelling in all metrics with an R of 0.811, RMSE of 1.324, MAE of 1.043, SD of 1.332, and CI of 0.813, with DAAP16 also delivering robust performance. For the CASF \(-\) 2013.195 test set, a similar trend is observed with our DAAP20 approach outperforming the nearest state-of-the-art predictor by a significant margin of 8%-20% across all evaluation metrics. The DAAP16 approach, not DAAP20, stands out on the ADS.74 dataset by surpassing predictors like Pafnucy, SFCNN and EGNA, showcasing substantial improvements of approximately 12%-37% in various metrics. When evaluating the CSAR-HiQ.51 and CSAR-HiQ.36 datasets against six state-of-the-art predictors, DAAP20 consistently outperforms all, indicating enhancements of 2%-20% and 3%-31%, respectively. Although DAAP16 does not surpass ResBiGAAT in CSAR-HiQ.51, it notably excels in the CSAR-HiQ.36 dataset, outperforming ResBiGAAT in all metrics except MAE. These results underscore the exceptional predictive capabilities of our DAAP approach across diverse datasets and evaluation criteria, consistently surpassing existing state-of-the-art predictors.
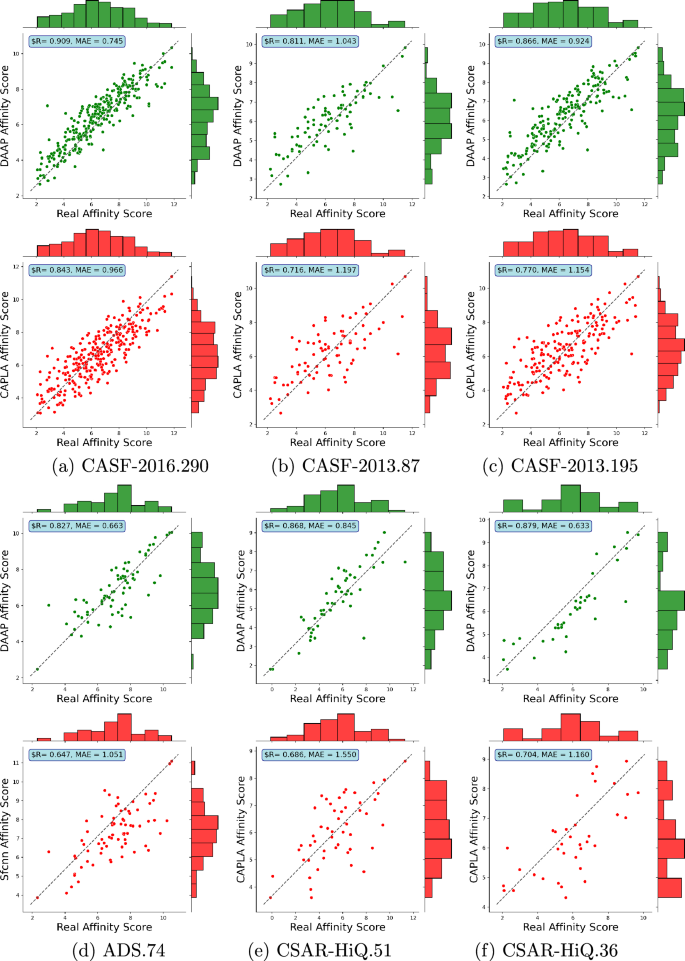
The distributions of real and predicted binding affinity values by our predictor (green) and the closest state-of-the-art predictor (red) across the six test sets
Figure 2 presents the distributions of actual and predicted binding affinities for our best DAAP approach and the closest state-of-the-art predictor. In all six test sets, a clear linear correlation and low mean absolute error (MAE) between predicted and actual binding affinity values can be observed for our DAAP model, demonstrating the strong performance of our model across these test sets. The other predictors show scattering over larger areas. In our analysis, we could not consider ResBiGAAT in the CSAR-HiQ.51 and CSAR-HiQ.36 datasets due to the unavailability of their results.
Ablation study and explainability
A significant contribution of this work is utilising distance matrix input features to capture critical information about the protein-ligand relationship. Specifically, we employ a concatenation of three distance maps, representing donor-acceptor, hydrophobic, and \(\pi \) -stacking interactions, as input features, effectively conveying essential protein-ligand bonding details. Following finalising our prediction architecture by incorporating two additional features derived from protein and SMILES sequences, we conduct an in-depth analysis of the impact of various combinations of these distance matrices as features. In the case of protein features, residues are selected based on which distance maps are considered.
Table 6 illustrates the outcomes obtained from experimenting with different combinations of distance maps and selected protein residue and ligand SMILES features on the CASF \(-\) 2016.290 test set. We devise four unique combinations, employing three distinct distance maps for both the PDBbind2016 and PDBbind2020 training datasets. Additionally, we explore a combination that integrates donor-acceptor, hydrophobic, and \(\pi \) -stacking distance maps with features from all protein residues, denoted as DA + \(\pi \) S + HP + FP, to evaluate the impact of using all residues versus selected ones.
From the information presented in Table 6 , it is evident that utilizing the donor-acceptor (DA) solely distance maps yields the lowest performance across both training sets, particularly when different combinations of distance maps are paired with selective protein residues. However, as expected, the combination of the three distance maps, namely DA, \(\pi \) S ( \(\pi \) -stacking), and HP (Hydrophobicity), demonstrates superior performance compared to other combinations. Notably, the combination of DA and HP outperforms the other two combinations but falls short of our best-performing feature set. The ensemble of DA, \(\pi \) S, HP and all protein residues exhibit the least favourable outcomes among the tested combinations. This result aligns with our expectations, as Hydrophobic interactions are the most prevalent in protein-ligand binding, underscoring their significance in feature analysis.
Integrating an attention mechanism into our model is crucial in achieving improved results. After consolidating the outputs of three 1D-CNN blocks, we employ attention, each receiving inputs from distance maps, protein sequences, and ligand sequences. The dimension of the feature is 384. As depicted in Fig. 3 , the heatmap visualization highlights the differential attention weights assigned to various features, with brighter and darker regions indicating higher weights to certain features, thus improving binding affinity predictions. This process underscores the mechanism’s ability to discern and elevate critical features, showing that not all features are equally important. Further emphasizing the significance of attention, a comparative analysis using the same model architecture without the attention mechanism on the same features-shown in the last row of Table 6 demonstrates its vital role in boosting predictive accuracy. This comparison not only reinforces the value of the attention mechanism in detecting intricate patterns within the feature space but also significantly enhances the model’s predictive capabilities.
Visualization of attention maps for concatenated features in the 1o0h protein-ligand complex of the CASF \(-\) 2016.290 dataset
Statistical analysis
In assessing the statistical significance of performance differences between DAAP and its closest competitors, Wilcoxon Signed Ranked Tests at a 95% confidence level were conducted. Comparisons included DAAP against CAPLA for CASF \(-\) 2016.290, CASF \(-\) 2013.87, CASF \(-\) 2013.195, CSAR-HiQ.36, and CSAR-HiQ.51 datasets and between DAAP and SFCNN for the ADS.74 test set. Unfortunately, ResBiGAAT’s results were unavailable for inclusion in the analysis. Table 7 depicts that DAAP demonstrated statistical significance compared to the closest state-of-the-art predictor across various test sets, as indicated by p-values ranging from 0.000 to 0.047. The consistently negative mean Z-values, ranging from \(-\) 14.71 to \(-\) 5.086, suggest a systematic improvement in predictive performance. Moreover, higher mean rankings, ranging from 19.5 to 144.5, further emphasize the overall superiority of DAAP. Notably, the superior performance is observed across diverse datasets, including CASF \(-\) 2016.290, CASF \(-\) 2013.87, CASF \(-\) 2013.195, ADS.74, CSAR-HiQ.51, and CSAR-HiQ.36. These findings underscore the robustness and effectiveness of DAAP in predicting protein-ligand binding affinity.
Screening results
In this section, we scrutinize the effectiveness of our predicted affinity scores to accurately differentiate between active binders (actives) and non-binders (decoys) throughout the screening procedure. To this end, we have carefully curated a subset of seven hand-verified targets from the Database of Useful Decoys: Enhanced (DUD-E), accessible via https://dude.docking.org , to serve as our evaluative benchmark. The details about seven targets are given in Table 8 . This table underscores the diversity and challenges inherent in the dataset, reflecting a wide range of D/A ratios that present a comprehensive framework for evaluating the discriminatory power of our predicted affinity scores.
To construct protein-ligand complexes for these targets, we employed AutoDock Vina, configuring the docking grid to a \(20\mathring{A} \times 20\mathring{A} \times 20\mathring{A}\) cube centred on the ligand’s position. This setup and 32 consecutive Monte-Carlo sampling iterations identified the optimal pose for each molecule pair. Our evaluation of the screening performance utilizes two pivotal metrics: the Receiver Operating Characteristic (ROC) curve [ 33 ] and the Enrichment Factor (EF) [ 34 ]. Figure 4 shows the ROC curve and the EF graph for a detailed examination of a predictive model’s efficacy in virtual screening. The ROC curve’s analysis, with AUC values spanning from 0.63 to 0.76 for the seven targets, illustrates our model’s proficient capability in differentiating between actives and decoys. These values, closely approaching the top-left corner of the graph, denote a high true positive rate alongside a low false positive rate, underscoring our model’s efficacy.
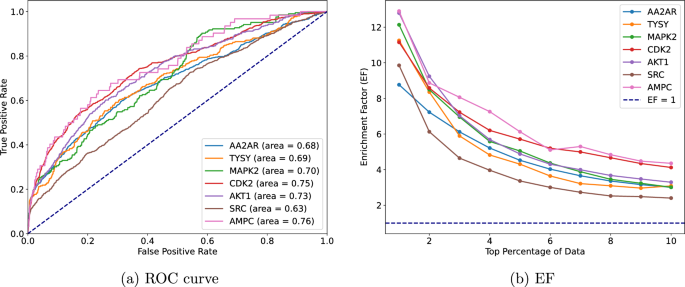
Screening Performance of the Predictive Model: Roc curve (left) and EF (right)
Furthermore, the EF graph of Fig. 4 provides a quantitative assessment of the model’s success in prioritizing active compounds within the top fractions of the dataset, notably the top 1% to 10%. Initial EF values ranging from 12.3 to 9.9 for the top 1% underscore our model’s exceptional ability to enrich active compounds beyond random chance significantly. This pronounced enrichment highlights the model’s utility in the early identification of promising candidates. However, the observed gradual decline in EF values with increasing dataset fractions aligns with expectations, reflecting the challenge of sustaining high enrichment levels across broader selections.
Conclusions
In our protein-ligand binding affinity prediction, we introduce atomic-level distance map features encompassing donor-acceptor, hydrophobic, and \(\pi \) -stacking interactions, providing deeper insights into interactions for precise predictions, both for short and long-range. We enhance our model further with specific protein sequence features of specific residues and ligand SMILES information. These features are integrated into an attention-based 1D-CNN architecture that is used a number of times for ensemble-based performance enhancement, resulting in superior results compared to existing methods across six benchmark datasets. Remarkably, on the CASF-2016 dataset, our model achieves a Correlation Coefficient (R) of 0.909, Root Mean Squared Error (RMSE) of 0.987, Mean Absolute Error (MAE) of 0.745, Standard Deviation (SD) of 0.988, and Concordance Index (CI) of 0.876, signifying its potential to advance drug discovery binding affinity prediction. The program and data are publicly available on the website https://gitlab.com/mahnewton/daap .
We describe the protein-ligand dataset used in our work. We also describe our proposed method in terms of its input features, output representations, and deep learning architectures.
Protein-ligand datasets
In the domain of protein-ligand binding affinity research, one of the primary sources for training, validation, and test sets is the widely recognized PDBbind database [ 35 ]. This database is meticulously curated. It comprises experimentally verified protein-ligand complexes. Each complex encompasses the three-dimensional structures of a protein-ligand pair alongside its corresponding binding affinities expressed as \(pK_d\) values. The PDBbind database ( http://www.pdbbind.org.cn/ ) is subdivided into two primary subsets: the general set and the refinement set . The PDBbind version 2016 dataset (named PDBbind2016) contains 9221 and 3685 unique protein-ligand complexes, while the PDBbind version 2020 dataset (named PDBbind2020) includes 14127 and 5316 protein-ligand complexes in the general and refinement sets, respectively.
Similar to the most recent state-of-the-art affinity predictors such as Pafnucy [ 20 ], DeepDTAF [ 23 ], OnionNet [ 3 ], DLSSAffinity [ 4 ], LuEtAl [ 36 ], EGNA [ 25 ] and CAPLA [ 26 ], our DAAP16 method is trained using the 9221 + 3685 = 12906 protein-ligand complexes in the general and refinement subsets of the PDBbind dataset version 2016 . Following the same training-validation set formation approach of the recent predictors such as Pafnucy, OnionNet, DeepDTAF, DLSSAffinity and CAPLA, we put 1000 randomly selected protein-ligand complexes in the validation set and the remaining 11906 distinct protein-ligand pairs in the training set. Another version of DAAP, named DAAP20, was generated using the PDBbind database version 2020 , which aligns with the training set of ResBiGAAT [ 27 ]. To avoid overlap, we filtered out protein-ligand complexes common between the PDBbind2020 training set and the six independent test sets. After this filtering process, 19027 unique protein-ligand complexes were retained for training from the initial pool of 19443 in PDBbind2020.
To ensure a rigorous and impartial assessment of the effectiveness of our proposed approach, we employ six well-established, independent blind test datasets. There is no overlap of protein-ligand complexes between the training sets and these six independent test sets.
CASF-2016.290 The 290 protein-ligand complexes, commonly referred to as CASF-2016, are selected from the PDBbind version 2016 core set ( http://www.pdbbind.org.cn/casf.php ) and have become the gold standard test set for recent affinity predictors such as DLSSAffinity [ 4 ], LuEtAl [ 36 ], EGNA [ 25 ] and CAPLA [ 26 ].
CASF-2013.87 and CASF-2013.195 Similar to the approach taken by DLSSAffinity [ 4 ], we carefully curated 87 unique protein-ligand complexes from the CASF-2013 dataset, which originally consists of 195 complexes ( http://www.pdbbind.org.cn/casf.php ). These 87 complexes were chosen to ensure no overlap with our training set or the CASF-2016 test set. Additionally, we use the entire set of 195 complexes as another test set, named CASF \(-\) 2013.195.
ADS.74 This test set from SFCNN [ 24 ] comprises 74 protein-ligand complexes sourced from the Astex diverse set [ 37 ].
CSAR-HiQ.51 and CSAR-HiQ.36 These two test datasets contain 51 and 36 protein-ligand complexes from the well-known CSAR [ 38 ] dataset. Recent affinity predictors such as EGNA [ 25 ], CAPLA and ResBiGAAT [ 26 , 27 ] have employed CSAR as a benchmark dataset. To get our two test datasets, we have followed the procedure of CAPLA and filtered out protein-ligand complexes with duplicate PDB IDs from two distinct CSAR subsets containing 176 and 167 protein-ligand complexes, respectively.
Input features
Given protein-ligand complexes in the datasets, we extract three distinctive features from proteins, ligands, and protein-ligand binding pockets. We describe these below.
Protein representation
We employ three distinct features for encoding protein sequences: one-hot encoding of amino acids, a Hidden Markov model based on multiple sequence alignment features (HHM), and seven physicochemical properties.
In the one-hot encoding scheme for the 20 standard amino acids and non-standard amino acids, each amino acid is represented by a 21-dimensional vector. This vector contains twenty “0 s” and one “1”, where the position of the “1” corresponds to the amino acid index in the protein sequence.
To construct the HHM features, we have run an iterative searching tool named HHblits [ 39 ] against the Uniclust30 database ( http://wwwuser.gwdg.de/~compbiol/uniclust/2020_06/ ) as of June 2020. This process allows us to generate HHM sequence profile features for the proteins in our analysis. Each resulting .hhm feature file contains 30 columns corresponding to various parameters such as emission frequencies, transition frequencies, and Multiple Sequence Alignment (MSA) diversities for each residue. Like EGNA, for columns 1 to 27, the numbers are transformed into frequencies using the formula \(f = 2^{-0.001*p}\) , where f represents the frequency, and p is the pseudo-count. This transformation allows the conversion of these parameters into frequency values. Columns 28 to 30 are normalized using the equation: \(f = \frac{0.001*p}{20}\) . This normalization process ensures that these columns are appropriately scaled for further analysis and interpretation.
The seven physicochemical properties [ 14 , 29 ] for each amino acid residue are steric parameter (graph shape index), hydrophobicity, volume, polarisability, isoelectric point, helix probability, and sheet probability. When extracting these three features for protein residues, we focused exclusively on the 20 standard amino acid residues. If a residue is non-standard, we assigned a feature value of 0.0.
In our approach, we initially concatenate all three features sequentially for the entire protein sequence. Subsequently, to enhance the specificity of our model, we employ a filtering strategy where residues lacking donor [ 40 ], hydrophobic [ 31 ], and \(\pi \) -stacking [ 32 ] atoms within their amino acid side chains are excluded from the analysis. Additionally, to prevent overlap, we select unique residues after identification based on donor, hydrophobic, or \(\pi \) -stacking atoms for each protein sequence. The rationale behind this filtering is to focus on residues that are actively involved in critical interactions relevant to protein-ligand binding. The resulting feature dimension for each retained protein residue is 58. This feature set includes one-hot encoding of amino acids, a Hidden Markov model based on multiple sequence alignment features (HHM), and seven physicochemical properties. These features are comprehensively summarised in Table 9 for clarity.
Considering the variable numbers of residues that proteins can possess, we have considered a standardized protein sequence length to align with the fixed-size requirements of deep learning algorithms. In our initial experiments exploring various sequence lengths in the datasets, we found that a maximum length of 500 yields better performance in terms of pearson correlation coefficient (R) and mean absolute error (MAE). If the number of selected residues falls below 500, we pad the sequence with zeros; conversely, if it exceeds 500, we truncate it to 500 from the initial position of the sequence. The final dimension of each protein is \(500\times 58\) .
Ligand representation
We use SMILES to represent ligands. SMILES is a widely adopted one-dimensional representation of chemical structures of ligands [ 41 ]. To convert ligand properties such as atoms, bonds, and rings from ligand SDF files into SMILES strings, we use the Open Babel chemical tool [ 42 ]. The SMILES strings comprise 64 unique characters, each corresponding to a specific numeric digit ranging from 1 to 64. For example, the SMILES string “HC(O=)N” is represented as [12, 42, 1, 48, 40, 31, 14]. In line with our protein representation approach, we set a fixed length of 150 characters for each SMILES string.
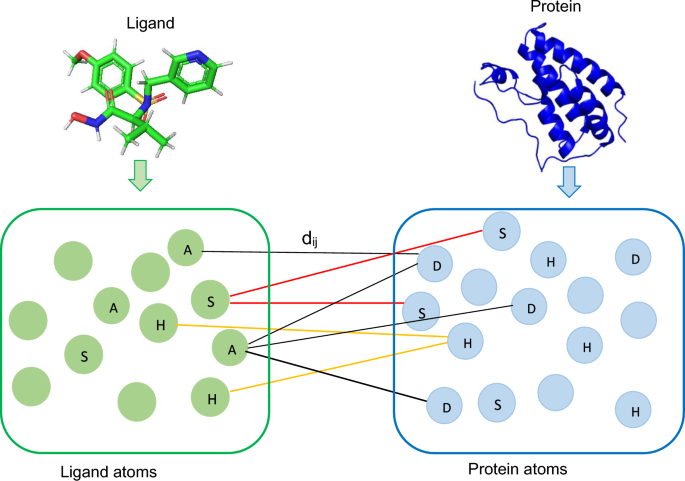
Various distance measures that potentially capture protein-ligand interactions. In the figure, \(d_{ij}\) represents the distance between a donor (D), hydrophobic (H), or \(\pi \) -stacking (S) atom i in the protein and the corresponding acceptor (A), hydrophobic (H), or \(\pi \) -stacking (S) atom j in the ligand. Empty circles represent other atom types. Different colour lines represent different types of interactions
Binding pocket representation
A binding pocket refers to a cavity located either on the surface or within the interior of a protein. A binding pocket possesses specific characteristics that make it suitable for binding a ligand [ 43 ]. Protein residues within the binding pocket region exert a direct influence, while residues outside this binding site can also have a far-reaching impact on affinity prediction. Among various protein-ligand interactions within the binding pocket regions, donor-acceptor atoms [ 30 ], hydrophobic contacts [ 31 , 32 ], and \(\pi \) -stacking [ 31 , 32 ] interactions are the most prevalent, and these interactions could significantly contribute to the enhancement of affinity score prediction. The formation of the protein-ligand complexes involves donor atoms from the proteins and acceptor atoms from the ligands. This process is subject to stringent chemical and geometric constraints associated with protein donor groups and ligand acceptors [ 30 ]. Hydrophobic interactions stand out as the primary driving force in protein-ligand interactions, while \(\pi \) -stacking interactions, particularly involving aromatic rings, play a substantial role in protein-ligand interactions [ 32 ]. However, there are instances where donor-acceptor interactions alone may not suffice, potentially failing to capture other interactions that do not conform to traditional donor-acceptor patterns. In such scenarios, hydrophobic contacts and \(\pi \) -stacking interactions become essential as they could provide valuable insights for accurate affinity prediction.
We employ three types of distance matrices in our work shown in Fig. 5 to capture protein-ligand interactions. The first one is the donor-acceptor distance matrix , which considers distances between protein donor atoms and acceptor ligand atoms, with data sourced from mol2/SDF files. We ensure that all ligand atoms contribute to the distance matrix construction, even in cases where ligands lack explicit acceptor atoms. Furthermore, we calculate the hydrophobic distance matrix by measuring the distance between hydrophobic protein atoms and hydrophobic ligand atoms, ensuring the distance is less than \(4.5\mathring{A}\) [ 31 ]. Similarly, we compute the \(\pi \) - stacking distance matrix by considering protein and ligand \(\pi \) -stacking atoms and applying a distance threshold of \(4.0\mathring{A}\) [ 32 ]. These three types of atoms are selected from the heavy atoms, referring to any atom that is not hydrogen.
We discretize the initially calculated real-valued distance matrices representing the three types of interactions into binned distance matrices. These matrices are constrained within a maximum distance threshold of \(20\mathring{A}\) . The decision to set a maximum distance threshold of \(20\mathring{A}\) for capturing the binding pocket’s spatial context is informed by practices in both affinity prediction and protein structure prediction fields. Notably, methodologies like Pafnucy [ 20 ], DLSSAffinity [ 4 ], and EGNA [ 25 ], as well as advanced protein structure prediction models such as AlphaFold [ 28 ] and trRosetta [ 44 ], utilize a 20Å range to define interaction spaces or predict structures. This consensus on the 20Å threshold reflects its sufficiency in providing valuable spatial information necessary for accurate modeling. The distance values ranging from \(0\mathring{A} - 20\mathring{A}\) are discretized into 40 bins, each with a \(0.5\mathring{A}\) interval. Any distance exceeding \(20\mathring{A}\) is assigned to the \(41^{st}\) bin. In our experimentation, we explored different distance ranges ( \(20\mathring{A}\) , \(25\mathring{A}\) , \(30\mathring{A}\) , \(35\mathring{A}\) , and \(40\mathring{A}\) ) while maintaining a uniform bin interval of \(0.5\mathring{A}\) . Among these ranges, \(20\mathring{A}\) yielded optimal results, and as such, we adopted it for our final analysis. Following this binning process, the original real-valued distances in the matrices are substituted with their corresponding bin numbers. Subsequently, we convert the 2D distance matrix into a 1D feature vector. We concatenate the three 1D vectors representing the three distinct interactions into a single vector to construct the final feature vector. To ensure consistency, the maximum length of the feature vector is set to 1000 for each pocket.
Output representations
This binding affinity is measured in the dissociation constant ( \(K_d\) ). For simplicity in calculations, the actual affinity score \(K_d\) is commonly converted into \(pK_d\) by taking the negative logarithm of \(K_d\) .
Deep learning architectures
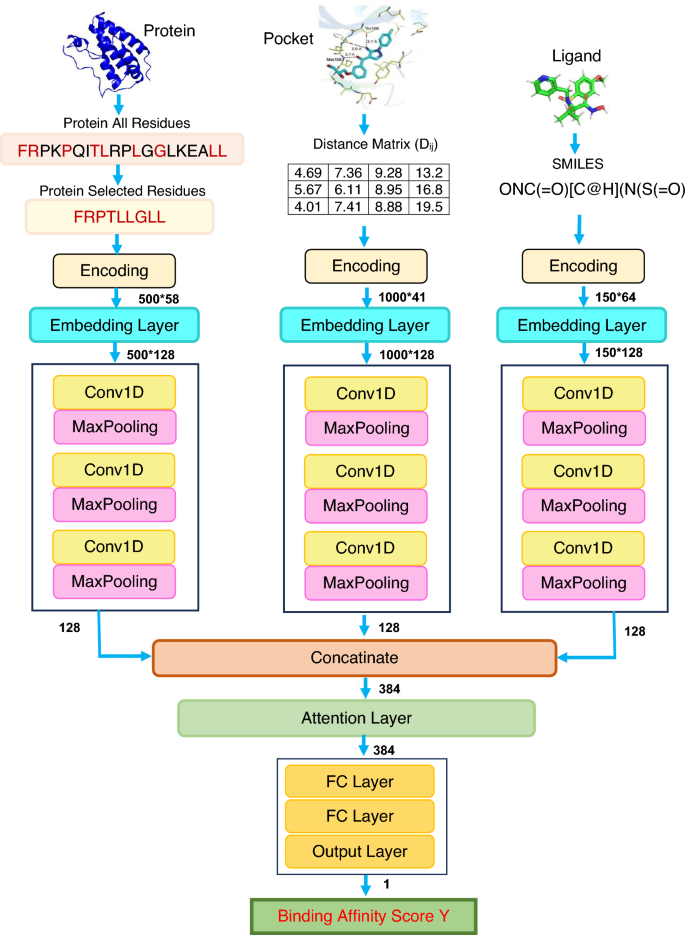
The proposed model architecture
We propose a deep-learning regression model to predict protein-ligand binding affinities, shown in Fig. 6 . Our model comprises three integral components: convolutional neural network (CNN), attention mechanism, and fully connected neural network (FCNN). Before feeding to the CNN block, information from three distinct feature sources (proteins, ligands, and interactions) is encoded and subsequently processed through the embedding layer. The embedding layer transforms the inputs into fixed-length vectors of a predefined size (in this case, 128 dimensions), enabling more effective feature representation with reduced dimensionality. During training, our model operates with a batch size of 16 and is optimized using the Adam optimizer and a learning rate set at 0.001. We adopt the log cosh loss function for this work to optimise the model’s performance. The training regimen consists of 200 epochs, with the best model selected based on the validation loss, and a dropout rate of 0.2 is applied. The explored hyperparameter settings are summarised in Table 10 . We have explored these settings, and after preliminary experiments, we have selected these values which are emboldened.
Convolutional neural network
Much like DLSSAffinity [ 4 ], our model employs three 1D-CNN blocks, each dedicated to processing distinct feature sources: proteins, ligands, and interactions in pockets. Each of these 1D-CNN blocks comprises three convolutional layers paired with three Maxpooling layers. The configuration of the first two 1D-CNN blocks includes 32, 64, and 128 filters, each with corresponding filter lengths of 4, 8, and 12. In contrast, the 1D-CNN block responsible for handling SMILES sequence inputs features filters with 4, 6, and 8 adjusted lengths. Each of the three 1D-CNN blocks in our model generates a 128-dimensional output. Subsequently, before progressing to the next stage, the outputs of these three 1D-CNN blocks are concatenated and condensed into a unified 384-dimensional output.
Attention mechanism
In affinity prediction, attention mechanisms serve as crucial components in neural networks, enabling models to allocate varying levels of focus to distinct facets of input data [ 5 ]. These mechanisms play a critical role in weighing the significance of different features or entities when assessing their interaction strength. The attention mechanism uses the formula below.
We use the Scaled Dot-Product Attention [ 45 ] mechanism to calculate and apply attention scores to the input data. The attention mechanism calculates query ( Q ), key ( K ), and value ( V ) matrices from the input data. In this context, Q is a vector capturing a specific aspect of the input, K represents the context or memory of the model with each key associated with a value, and V signifies the values linked to the keys. It computes attention scores using the dot product of Q and K matrices, scaled by the square root of the dimensionality ( \(d_k\) ). Subsequently, a softmax function normalises the attention scores. Finally, the output is generated as a weighted summation of the value (V) matrix, guided by the computed attention scores.
Notably, the output of the concatenation layer passes through the attention layer. The input to the attention layer originates from the output of the concatenation layer, preserving the same dimensionality as the input data. This design ensures the retention of crucial structural information throughout the attention mechanism.
Fully connected neural network
The output of the attention layer transitions into the subsequent stage within our model architecture, known as the Fully Connected Neural Network (FCNN) block. The FCNN block consists of two fully connected (FC) layers, where the two layers have 256 and 128 nodes respectively. The final stage in our proposed prediction model is the output layer, which follows the last FC layer.
Evaluation metrics
We comprehensively evaluate our affinity prediction model using five well-established performance metrics. The Pearson Correlation Coefficient (R) [ 4 , 24 , 26 , 36 ] measures the linear relationship between predicted and actual values. The Root Mean Square Error (RMSE) [ 4 , 24 , 26 ] and the Mean Absolute Error (MAE) [ 24 , 26 ] assess prediction accuracy and error dispersion. The Standard Deviation (SD) [ 4 , 24 , 26 , 36 ] evaluates prediction consistency, and the Concordance Index (CI) [ 26 , 36 ] determines the model’s ability to rank protein-ligand complexes accurately. Higher R and CI values and lower RMSE, MAE, and SD values indicate better prediction accuracy. These metrics are collectively very robust measures for comparison of our model’s performance against that of the state-of-the-art techniques in the field of affinity prediction.
N : the number of protein-ligand complexes
\(Y_{\text {act}}\) : experimentally measured actual binding affinity values for the protein-ligand complexes
\(Y_{\text {pred}}\) : the predicted binding affinity values for the given protein-ligand complexes
\(y_{\text {act}_i}\) and \(y_{\text {pred}_i}\) : respectively the actual and predicted binding affinity value of the \(i^{th}\) protein-ligand complex
a : is slope
b : interpretation of the linear regression line of the predicted and actual values. Z : the normalization constant, i.e. the number of data pairs with different label values.
h ( u ): the step function that returns 1.0, 0.5, and 0.0for \(u>0\) , \(u = 0\) , and \(u<0\) respectively.
Availability of data and materials
The program and corresponding data are publicly available on the website https://gitlab.com/mahnewton/daap .
DiMasi JA, Grabowski HG, Hansen RW (2016) Innovation in the pharmaceutical industry: new estimates of r &d costs. J Health Econ 47:20–33
Article PubMed Google Scholar
Gilson MK, Zhou H-X (2007) Calculation of protein-ligand binding affinities. Ann Rev Biophys Biomol Str 36(1):21–42
Article CAS Google Scholar
Zheng L, Fan J, Mu Y (2019) Onionnet: a multiple-layer intermolecular-contact-based convolutional neural network for protein-ligand binding affinity prediction. ACS Omega 4(14):15956–15965
Article CAS PubMed PubMed Central Google Scholar
Wang H, Liu H, Ning S, Zeng C, Zhao Y (2022) Dlssaffinity: protein-ligand binding affinity prediction via a deep learning model. Phys Chem Chem Phys 24(17):10124–10133
Article CAS PubMed Google Scholar
Seo S, Choi J, Park S, Ahn J (2021) Binding affinity prediction for protein-ligand complex using deep attention mechanism based on intermolecular interactions. BMC Bioinform 22(1):1–15
Article Google Scholar
Deng W, Breneman C, Embrechts MJ (2004) Predicting protein- ligand binding affinities using novel geometrical descriptors and machine-learning methods. J Chem Inf Comput Sci 44(2):699–703
Li L, Wang B, Meroueh SO (2011) Support vector regression scoring of receptor-ligand complexes for rank-ordering and virtual screening of chemical libraries. J Chem Inf Modeling 51(9):2132–2138
Ballester PJ, Mitchell JB (2010) A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics 26(9):1169–1175
Li H, Peng J, Sidorov P, Leung Y, Leung K-S, Wong M-H, Lu G, Ballester PJ (2019) Classical scoring functions for docking are unable to exploit large volumes of structural and interaction data. Bioinformatics 35(20):3989–3995
Deng L, Platt J. Ensemble deep learning for speech recognition. In: Proc. Interspeech. 2014
Chen C, Seff A, Kornhauser A, Xiao J. Deepdriving: learning affordance for direct perception in autonomous driving. In: Proceedings of the IEEE International Conference on Computer Vision, 2015; pp. 2722–2730
Lin T-Y, RoyChowdhury A, Maji S (2017) Bilinear convolutional neural networks for fine-grained visual recognition. IEEE Trans Pattern Anal Mach Intell 40(6):1309–1322
Newton MH, Rahman J, Zaman R, Sattar A. Enhancing protein contact map prediction accuracy via ensembles of inter-residue distance predictors. Computational Biology and Chemistry, 2022; 107700.
Rahman J, Newton MH, Hasan MAM, Sattar A (2022) A stacked meta-ensemble for protein inter-residue distance prediction. Comput Biol Med 148:105824
Isert C, Atz K, Schneider G (2023) Structure-based drug design with geometric deep learning. Curr Opin Struct Biol 79:102548
Krentzel D, Shorte SL, Zimmer C (2023) Deep learning in image-based phenotypic drug discovery. Trend Cell Biol 33(7):538–554
Yang L, Jin C, Yang G, Bing Z, Huang L, Niu Y, Yang L (2023) Transformer-based deep learning method for optimizing admet properties of lead compounds. Phys Chem Chem Phys 25(3):2377–2385
Masters MR, Mahmoud AH, Wei Y, Lill MA (2023) Deep learning model for efficient protein-ligand docking with implicit side-chain flexibility. J Chem Inf Modeling 63(6):1695–1707
Öztürk H, Özgür A, Ozkirimli E (2018) Deepdta: deep drug-target binding affinity prediction. Bioinformatics 34(17):821–829
Stepniewska-Dziubinska MM, Zielenkiewicz P, Siedlecki P (2018) Development and evaluation of a deep learning model for protein-ligand binding affinity prediction. Bioinformatics 34(21):3666–3674
Jiménez J, Skalic M, Martinez-Rosell G, De Fabritiis G (2018) \(k_{deep}\) : protein-ligand absolute binding affinity prediction via 3d-convolutional neural networks. J Chem Inf Modeling 58(2):287–296
Li Y, Rezaei MA, Li C, Li X (2019) Deepatom: a framework for protein-ligand binding affinity prediction. In: 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 303–310, IEEE
Wang K, Zhou R, Li Y, Li M (2021) Deepdtaf: a deep learning method to predict protein-ligand binding affinity. Brief Bioinf 22(5):072
Wang Y, Wei Z, Xi L (2022) Sfcnn: a novel scoring function based on 3d convolutional neural network for accurate and stable protein-ligand affinity prediction. BMC Bioinform 23(1):1–18
Xia C, Feng S-H, Xia Y, Pan X, Shen H-B (2023) Leveraging scaffold information to predict protein-ligand binding affinity with an empirical graph neural network. Brief Bioinf. https://doi.org/10.1093/bib/bbac603
Jin Z, Wu T, Chen T, Pan D, Wang X, Xie J, Quan L, Lyu Q (2023) Capla: improved prediction of protein-ligand binding affinity by a deep learning approach based on a cross-attention mechanism. Bioinformatics 39(2):049
Abdelkader GA, Njimbouom SN, Oh T-J, Kim J-D (2023) Resbigaat: Residual bi-gru with attention for protein-ligand binding affinity prediction. Computational Biology and Chemistry, 107969
Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AW, Bridgland A et al (2020) Improved protein structure prediction using potentials from deep learning. Nature 577(7792):706–710
Rahman J, Newton MH, Islam MKB, Sattar A (2022) Enhancing protein inter-residue real distance prediction by scrutinising deep learning models. Sci Rep 12(1):787
Raschka S, Wolf AJ, Bemister-Buffington J, Kuhn LA (2018) Protein-ligand interfaces are polarized: discovery of a strong trend for intermolecular hydrogen bonds to favor donors on the protein side with implications for predicting and designing ligand complexes. J Computer-aided Mol Design 32:511–528
Jubb HC, Higueruelo AP, Ochoa-Montaño B, Pitt WR, Ascher DB, Blundell TL (2017) Arpeggio: a web server for calculating and visualising interatomic interactions in protein structures. J Mol Biol 429(3):365–371
Freitas RF, Schapira M (2017) A systematic analysis of atomic protein-ligand interactions in the pdb. Medchemcomm 8(10):1970–1981
Empereur-Mot C, Guillemain H, Latouche A, Zagury J-F, Viallon V, Montes M (2015) Predictiveness curves in virtual screening. J Cheminf 7(1):1–17
Li H, Zhang H, Zheng M, Luo J, Kang L, Liu X, Wang X, Jiang H (2009) An effective docking strategy for virtual screening based on multi-objective optimization algorithm. BMC Bioinf 10:1–12
Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK (2007) Bindingdb: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res 35(suppl–1):198–201
Lu Y, Liu J, Jiang T, Guan S, Wu H. Protein-ligand binding affinity prediction based on deep learning. In: International Conference on Intelligent Computing, 2022; pp. 310–316. Springer.
Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WT, Mortenson PN, Murray CW (2007) Diverse, high-quality test set for the validation of protein- ligand docking performance. J Med Chem 50(4):726–741
Dunbar JB Jr, Smith RD, Yang C-Y, Ung PM-U, Lexa KW, Khazanov NA, Stuckey JA, Wang S, Carlson HA (2011) Csar benchmark exercise of 2010: selection of the protein-ligand complexes. J Chem Inf Modeling 51(9):2036–2046
Remmert M, Biegert A, Hauser A, Söding J (2012) Hhblits: lightning-fast iterative protein sequence searching by hmm-hmm alignment. Nat Methods 9(2):173–175
Hydrogen donor and acceptor atoms of the amino acid. https://www.imgt.org/IMGTeducation/Aide-memoire/_UK/aminoacids/charge/ . Accessed: 13-08-2023
Weininger D (1988) Smiles, a chemical language and information system. 1. introduction to methodology and encoding rules. J Chem Inf Comput Sci 28(1):31–36
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open babel: An open chemical toolbox. J Cheminf 3(1):1–14
Google Scholar
Stank A, Kokh DB, Fuller JC, Wade RC (2016) Protein binding pocket dynamics. Accounts Chem Res 49(5):809–815
Yang J, Anishchenko I, Park H, Peng Z, Ovchinnikov S, Baker D (2020) Improved protein structure prediction using predicted interresidue orientations. Proc Natl Acad Sci 117(3):1496–1503
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Advances in neural information processing systems 30
Download references
Acknowledgements
This research is partially supported by the research seed grant awarded to M.A.H.N. at the University of Newcastle. The research team acknowledges the valuable assistance of the Griffith University eResearch Service & Specialised Platforms team for granting access to their High-Performance Computing Cluster, which played a crucial role in completing this research endeavour.
This research is partially supported by the research seed Grant awarded to M.A.H.N. at the University of Newcastle.
Author information
Julia Rahman and M. A. Hakim Newton are co-first-authors and contributed equally.
Authors and Affiliations
School of Information and Communication Technology, Griffith University, 170 Kessels Rd, Nathan, 4111, QLD, Australia
Julia Rahman
Institute for Integrated and Intelligent Systems (IIIS), Griffith University, 170 Kessels Rd, Nathan, 4111, QLD, Australia
M. A. Hakim Newton & Abdul Sattar
School of Information and Physical Sciences, University of Newcastle, University Dr, Callaghan, 2308, NSW, Australia
M. A. Hakim Newton
Department of Computer Science & Engineering, Bangladesh University of Engineering and Technology, Palashi, 1205, Dhaka, Bangladesh
Mohammed Eunus Ali
You can also search for this author in PubMed Google Scholar
Contributions
The contributions of the authors to this work were as follows: J.R. and M.A.H.N. played equal roles in all aspects of the project, including conceptualization, data curation, formal analysis, methodology, software development, and writing of the initial draft. M.E.A. helped in the concept development, review and editing of the manuscript. A.S. actively engaged in discussions, facilitated funding acquisition, provided supervision, and thoroughly reviewed the manuscript.
Corresponding author
Correspondence to Julia Rahman .
Ethics declarations
Competing interests.
No Conflict of interest is declared.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Rahman, J., Newton, M.A.H., Ali, M.E. et al. Distance plus attention for binding affinity prediction. J Cheminform 16 , 52 (2024). https://doi.org/10.1186/s13321-024-00844-x
Download citation
Received : 01 December 2023
Accepted : 24 April 2024
Published : 12 May 2024
DOI : https://doi.org/10.1186/s13321-024-00844-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Binding affinity
- Distance matrix
- Donor-acceptor
- Hydrophobicity
- \(\pi \) -Stacking
- Deep learning
Journal of Cheminformatics
ISSN: 1758-2946
- Submission enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
A class of methods known as the T-Method has recently been developed for use in forecasting. The T-Method is a subset of the Taguchi System of Quality Engineering. The T-Method is used to calculate the S/N ratio associated with the overall prediction of a data set. Many forecasting models exist in the literature.
The purpose of prediction is to make informed decisions and take actions based on expected future outcomes. Predictions are used to estimate the likelihood of future events or outcomes, and to guide decision-making based on those estimates. In many industries and fields, predictions are an essential tool for optimizing resources, managing risks ...
This thesis examines the application of machine learning algorithms to predict whether a student will be successful or not. The specific focus of the thesis is the comparison of machine learning methods and feature engineering techniques in terms of how much they improve the prediction performance.
This thesis introduces novel methods for producing well-calibrated probabilistic predictions for machine learning classification and regression problems. A new method for multi-class ...
Validation of prediction models is highly recommended and increasingly common in the literature. A systematic review of validation studies is therefore helpful, with meta-analysis needed to summarise the predictive performance of the model being validated across different settings and populations. This article provides guidance for researchers systematically reviewing and meta-analysing the ...
Step 1: Explain your methodological approach. Step 2: Describe your data collection methods. Step 3: Describe your analysis method. Step 4: Evaluate and justify the methodological choices you made. Tips for writing a strong methodology chapter. Other interesting articles.
Bournemouth. [email protected]. Abstract —The purpose of this study is to review time series forecasting methods and briefly explains the working of time series. forecasting methods. We ...
2.6 Machine Learning in Crop Yield Prediction. Throughout the recent years, machine learning has been experimented in order to make. yield forecasts. A recent research study by Kamath et al. (2021) was conducted to. forecast the yield of major crops in India. They used the classification algorithm Random.
based methods such as poor template quality or generality. Typically, these methods represent ... thesis and forward reaction prediction. The retrosynthesis prediction problem has two scenarios: known and unknown reaction class. In the known reaction class case, a class identifier is appended to the beginning of the target molecule, ...
In this thesis, we develop a collection of state-of-the-art deep learning models for time series forecasting. Primarily focusing on a closer alignment with traditional methods in time series modelling, we adopt three main directions of research -- 1) novel architectures, 2) hybrid models, and 3) feature extraction.
Evaluation of machine learning regression methods is based on the degree of agreement between predicted and observed values. The RMSE, R 2, and MAE statistical measures check on the precision of a prediction or forecasting model. Machine learning excels at rainfall prediction regardless of climate or timescale.
The existing models can be broadly categorized into two groups: mechanistic water quality prediction methods and non-mechanistic water quality prediction methods. Next, each of the two types of methods involved in the existing literature is analyzed and presented, and Fig. 1 shows the classification strategy and the linear structure of this paper.
Two Final Tips: When you're writing your justification, write for your audience. Your purpose here is to provide more than a technical list of details and procedures. This section should focus more on the why and less on the how. Consider your methodology as you're conducting your research.
A theory of personality should strive to predict humans' thoughts, feelings, and behaviors across different life contexts. Indeed, the representation we discovered in Study 1 was superior to the ...
traditional churn analysis and prediction methods that focus on classifying customer expected to churn in a predefined time window, this thesis project draws analogies with the study of reliability and maintenance engineering in order to quantitatively compare effectivity of customer-specific retention investments.
Summary "The Lockwood Analytical Method for Prediction (LAMP) is a systematic technique for predicting short-term, unique behaviors. Using primarily qualitative empirical data, LAMP allows the analyst to predict the most likely outcomes for specific research questions across a wide range of intelligence problems, such as cyber threats in the U.S., the possibility of an Al Qaeda attack, the ...
Accurate wave height prediction is significant in ports, energy, fisheries, and other offshore operations. In this study, a regional significant wave height prediction model with a high spatial and temporal resolution is proposed based on the ConvLSTM algorithm. The model learns the intrinsic correlations of the data generated by the numerical model, making it possible to combine the ...
predicted (solid) using the third prediction method in Well 1 (1-300 testing data). ..... 59 Figure 4.5 Comparison of reduction in prediction performance when one of the 15 conventional logs is removed one at a time.
[1] Gao Z., Gu Y., 5G traffic prediction based on deep learning, in: Computational Intelligence and Neuroscience, 2022, pp. 1687 - 5265, 10.1155/2022/3174530. Google Scholar Digital Library [2] Liang X., Research on network security filtering model and key algorithms based on network abnormal traffic analysis, in: 2021 International Conference on Networking, Communications and Information ...
Protein structure prediction is an interdisciplinary research topic that has attracted researchers from multiple fields, including biochemistry, medicine, physics, mathematics, and computer science. These researchers adopt various research paradigms to attack the same structure prediction problem: biochemists and physicists attempt to reveal the principles governing protein folding ...
We developed pooled and individualized predictors using the following methods: (1) iv support vector machines (SVM); and (2) multilayer perceptrons (MLP). We then assessed model performance using epileptic rat data. MLP yielded the highest AUROC of 0.88 on our pooled dataset of 1012 rodent seizures.
The proposed LSTM-based diabetes prediction algorithm is trained with 80% of the data, and the remaining 20% is used for testing. We fine-tuned the prediction model by using a different number of LSTM units in the cell state. This fine-tuning helps to identify more prominent features in the dataset.
Deep learning-based weather prediction (DLWP) is expected to be a strong supplement to the conventional method. At present, many researchers have tried to introduce data-driven deep learning into ...
The proposed approaches have been validated on both synthetic and real data, and compared against other methods from the literature. It results that neighborhood analysis allows to outperform competitors, and when it is combined with collaborative filtering the prediction accuracy further improves, scoring a value of AUC equal to 0966.
A machine learning (ML) method is generalizable if it can make predictions on inputs which differ from the training dataset. For predictions of wave-induced ship responses, generalizability is an important consideration if ML methods are to be useful in design evaluations. Furthermore, the size of the training dataset has a significant impact on the practicality of a method, especially when ...
The work in this dissertation tries to further the quest for a world where everyone is aware of uncertainty, of how important it is and how to embrace it instead of fearing it. A specific, though general, framework that allows anyone to obtain accurate uncertainty estimates is singled out and analysed. ... conformal prediction is, at the time ...
What Makes a Grade 7 Essay on An Inspector Calls Quotes 9 Explanations 21 NAMED METHODS 3 Context 10 Society 2 Patriarchal 0 Thesis Statement Y Conclusion Y Words 643 You are probably wondering what an explanation is. It is anything which means: this implies, indicates, suggests, connotes, Priestley wants us to think, feel, believe, understand..
Inspired by the use of distance measures in protein structure prediction [ 14, 28, 29 ], in this work, we employ distance-based input features in protein-ligand binding affinity prediction. To be more specific, we use distances between donor-acceptor [ 30 ], hydrophobic [ 31, 32 ], and π -stacking [ 31, 32] atoms as interactions between such ...
Research on vehicle trajectory prediction based on road monitoring video data often utilizes a global map as an input, disregarding the fact that drivers rely on the road structures observable from their own positions for path planning. This oversight reduces the accuracy of prediction. To address this, we propose the CVAE-VGAE model, a novel trajectory prediction approach.
The aim of this report is to conduct a comparative study on the most commonly used Time Series estimators in order to benchmark their performance on a wide variety of series from different fields ...